miRNA biomarker signatures for Multiple Sclerosis disease states, Sci. Rep. 2017
That’s not the only way by which a biomarker, contained in exosomes, can be used to assess disease states.
Other than proteins, another major component we can find in exosomes is microRNA (miRNA). A recent paper assessed exosomal miRNA biomarkers and signatures in multiple sclerosis in order to determine if they could accurately reflect a patient’s disease status.
As you might be aware, multiple sclerosis (MS) is a chronic inflammatory, demyelinating, neurodegenerative disorder.
The clinical phenotypes of MS are numerous and include the following:
- Relapsing-remitting MS (RRMS)
- Primary progressive MS (PPMS)
- Secondary progressive MS (SPMS).
While RRMS is the most prevalent form of MS, comprising over 70% of all cases, within 10-15 years of disease onset the majority of patients with RRMS will progress to SPMS. Unfortunately, SPMS is a phase of MS that is known for its gradual worsening of clinical signs and symptoms and a heterogeneous or complete lack of response to existing therapy.
As of now, there’s no single definitive test for MS. Therefore, it can be difficult to diagnose this disorder as its diagnosis and the monitoring of disease progression relies on multiple (sometimes subjective) clinical parameters; such as physical exams, MRI, and assessments of CSF.
Ongoing research indicates that circulating exosomes are promising candidate biomarkers for MS since they may contain everything from nucleic acids to proteins and can cross the blood-brain barrier. Biomarkers that can be used to identify patients who are able to benefit from therapy the most.
In other words, there’s a big need for objective, easy, even minimally invasive liquid biopsies and markers (like miRNA biomarkers) that can stratify patients with MS.
Consequently, the authors of one study hypothesized that serum exosomal miRNAs could represent a useful blood-based assay to detect and monitor MS.
The researchers started out by isolating and characterizing extracellular vesicles (EVs) derived from the serum of controls and patients with MS.
Using 1 mL of serum from each individual, the authors treated the serum samples with RNaseA in order to remove any unprotected circulating RNA, isolated the exosomes using size exclusion chromatography, and analyzed them via nanoparticle tracking analysis and transmission electron microscopy.
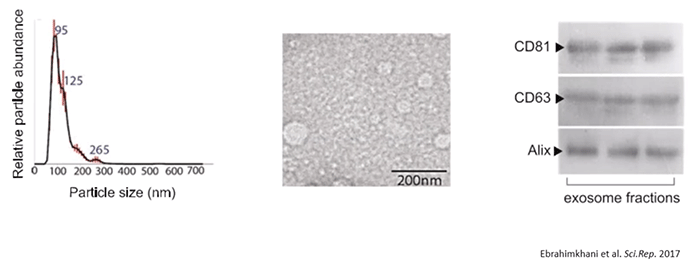
Diagram 2.6: Identification & Characterization of Serum-Derived EVs.
The presence of exosomal markers like CD81, CD63, and Alix was confirmed (Diagram 2.6).
RNA extraction yielded an RNA profile we’d expect for exosomes; where the bioanalyzer showed a typical peak between 25-200 nt without any indication of ribosomal RNA. The authors also compared miRNA profiles between samples with and without RNAse pre-treatment (Diagram 2.7).
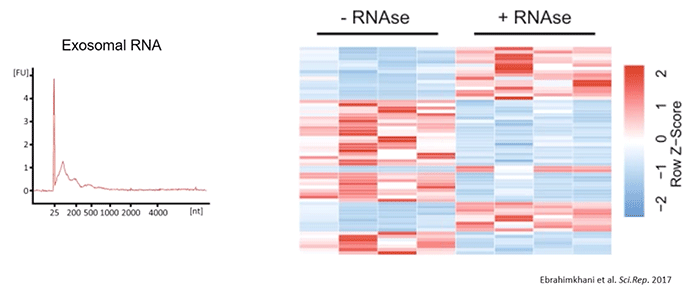
Diagram 2.7: Serum Exosomes Carry Unique miRNA Signatures.
The researchers were able to identify a set of exosomal miRNAs that were differentially expressed between subtypes of MS.
Furthermore, they assessed receiver operator characteristic (ROC) curves and AUC measures for each candidate miRNA; for both RRMS and S/PPMS as compared to control. By combining different sets of miRNAs, the researchers created miRNA signatures that improved the ability to distinguish between subtypes of MS and control.
And while numerous candidate miRNAs have been proposed as potential biomarkers for MS in the past, the biomarkers identified in this study were, by and large, completely novel. This might be because of various differences between the unique constituent profiles of exosomes as compared to free circulating miRNAs.
Most importantly, this research showed that serum exosomes are a novel source of potentially useful miRNA biomarkers.
Download the Ebook
Click here to download the pdf version of the ebook