Rat SeraMir Exosome RNA Profiling Kit
Products
| Catalog Number | Description | Size | Price | Quantity | Add to Cart | |||
|---|---|---|---|---|---|---|---|---|
| RA812A-1 | Rat SeraMir Exosome RNA 384 microRNA qPCR Profiler Assay Set | 20 Profiles | $696 |
|
||||
Overview
Overview
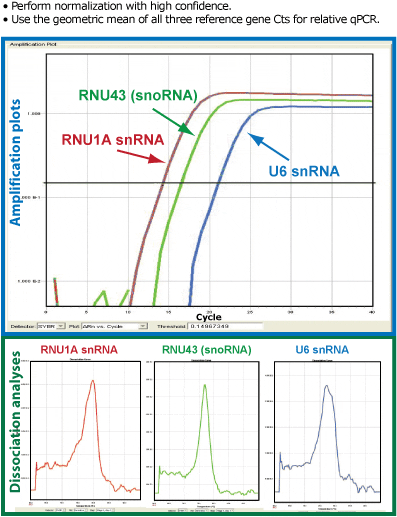
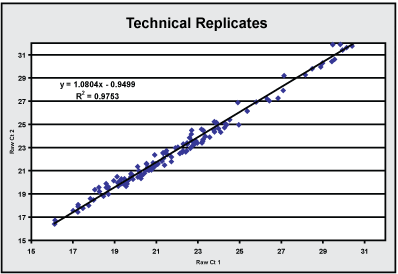
Quickly find out which miRNAs are packaged in your exosomes If you’ve already extracted, amplified, and tagged miRNAs from exosomes using SeraMir and are ready for profiling, turn to our Rat SeraMir Exosome RNA Profiling Kit. This kit enables the generation of up to twenty profiles against 380 rat miRNAs, and includes control wells for robust quantitation (find out which miRNAs are included by clicking here). Choose the SeraMir Kit that’s right for you| Cat. # | Name | Kit includes | |||
|---|---|---|---|---|---|
| ExoQuick or ExoQuick-TC | SeraMir RNA columns and reagents | SeraMir cDNA synthesis and amplification reagents | 384-well plate miRNAs for human, mouse, or rat | ||
| RA800A-1 | Complete SeraMir Exosome RNA Amplification Kit | ||||
| RA800TC-1 | Complete SeraMir Exosome RNA Amplification Kit for Media and Urine | ||||
| RA806A-1 | SeraMir Exosome RNA Purification Kit | ||||
| RA806TC-1 | SeraMir Exosome RNA Purification Kit for Media and Urine | ||||
| RA808A-1 | SeraMir Exosome RNA Purification Column Kit | ||||
| RA820A-1 | Human Complete SeraMir Exosome RNA Amplification and Profiling Kit | ||||
| RA820TC-1 | Human Complete SeraMir Exosome RNA Amplification and Profiling Kit for Media and Urine | ||||
| RA821A-1 | Mouse Complete SeraMir Exosome RNA Amplification and Profiling Kit | ||||
| RA821TC-1 | Mouse Complete SeraMir Exosome RNA Amplification and Profiling Kit for Media and Urine | ||||
| RA822A-1 | Rat Complete SeraMir Exosome RNA Amplification and Profiling Kit | ||||
| RA822TC-1 | Rat Complete SeraMir Exosome RNA Amplification and Profiling Kit for Media and Urine | ||||
References
How It Works
Supporting Data
Supporting Data
Better qPCR profiling with SeraMir
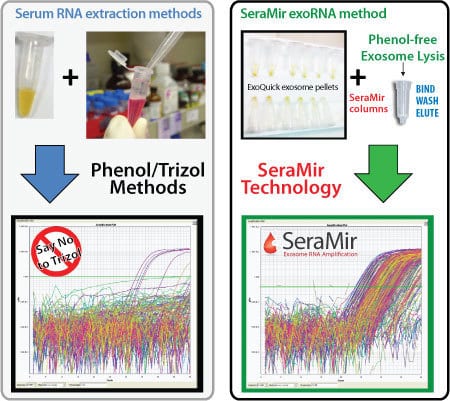
Figure 1. Serum RNA prepared by the SeraMir Kit delivers more reliable, reproducible qPCR profiles than when the RNA is isolated using conventional Trizol methods. Profiling of 380 Human microRNAs across the SeraMir 384 Profiler. The phenol-free exosome lysis step coupled to the small RNA binding columns isolates exoRNAs with much higher purity than Trizol/Phenol based methods. The SeraMir exoRNAs are compatible with downstream polyadenylation and reverse trancription reactions for amplification and accurate qPCR profiling.
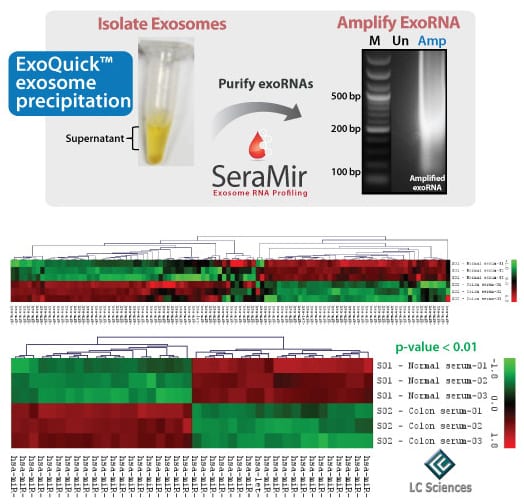
Figure 2. Serum exoRNAs prepared using SeraMir deliver excellent performance in microarray studies. Samples from a pooled normal serum preparation and from a male caucasian (age 73) with adenocarcinoma of the colon were used in this study. Exosomes were precipitated from 250 µL of serum using the SeraMir Exosome RNA Amplification Kit. The T7-amplified “sense” exoRNAs were then used for direct labeling analyses on LC Sciences miRBase ver.16 array chips (performed in triplicate). The exoRNAs were hybridized across 1,214 different microRNAs on the probe set.
Of the 1,214 microRNAs analyzed, 79 microRNAs showed a signal intensity >32. Within this set of 79, there was a clear colon versus normal “signature set” of 40 microRNAs that could discriminate normal from colon cancer serum samples with a p-value < 0.01. The identities of the microRNAs found in this study have been masked while further investigation continues.
FAQs
Documentation
Citations
Related Products
Products
| Catalog Number | Description | Size | Price | Quantity | Add to Cart | |||
|---|---|---|---|---|---|---|---|---|
| RA812A-1 | Rat SeraMir Exosome RNA 384 microRNA qPCR Profiler Assay Set | 20 Profiles | $696 |
|
||||
Overview
Overview
Quickly find out which miRNAs are packaged in your exosomes If you’ve already extracted, amplified, and tagged miRNAs from exosomes using SeraMir and are ready for profiling, turn to our Rat SeraMir Exosome RNA Profiling Kit. This kit enables the generation of up to twenty profiles against 380 rat miRNAs, and includes control wells for robust quantitation (find out which miRNAs are included by clicking here). Choose the SeraMir Kit that’s right for you| Cat. # | Name | Kit includes | |||
|---|---|---|---|---|---|
| ExoQuick or ExoQuick-TC | SeraMir RNA columns and reagents | SeraMir cDNA synthesis and amplification reagents | 384-well plate miRNAs for human, mouse, or rat | ||
| RA800A-1 | Complete SeraMir Exosome RNA Amplification Kit | ||||
| RA800TC-1 | Complete SeraMir Exosome RNA Amplification Kit for Media and Urine | ||||
| RA806A-1 | SeraMir Exosome RNA Purification Kit | ||||
| RA806TC-1 | SeraMir Exosome RNA Purification Kit for Media and Urine | ||||
| RA808A-1 | SeraMir Exosome RNA Purification Column Kit | ||||
| RA820A-1 | Human Complete SeraMir Exosome RNA Amplification and Profiling Kit | ||||
| RA820TC-1 | Human Complete SeraMir Exosome RNA Amplification and Profiling Kit for Media and Urine | ||||
| RA821A-1 | Mouse Complete SeraMir Exosome RNA Amplification and Profiling Kit | ||||
| RA821TC-1 | Mouse Complete SeraMir Exosome RNA Amplification and Profiling Kit for Media and Urine | ||||
| RA822A-1 | Rat Complete SeraMir Exosome RNA Amplification and Profiling Kit | ||||
| RA822TC-1 | Rat Complete SeraMir Exosome RNA Amplification and Profiling Kit for Media and Urine | ||||
References
Supporting Data
Supporting Data
Better qPCR profiling with SeraMir
Figure 1. Serum RNA prepared by the SeraMir Kit delivers more reliable, reproducible qPCR profiles than when the RNA is isolated using conventional Trizol methods. Profiling of 380 Human microRNAs across the SeraMir 384 Profiler. The phenol-free exosome lysis step coupled to the small RNA binding columns isolates exoRNAs with much higher purity than Trizol/Phenol based methods. The SeraMir exoRNAs are compatible with downstream polyadenylation and reverse trancription reactions for amplification and accurate qPCR profiling.
Figure 2. Serum exoRNAs prepared using SeraMir deliver excellent performance in microarray studies. Samples from a pooled normal serum preparation and from a male caucasian (age 73) with adenocarcinoma of the colon were used in this study. Exosomes were precipitated from 250 µL of serum using the SeraMir Exosome RNA Amplification Kit. The T7-amplified “sense” exoRNAs were then used for direct labeling analyses on LC Sciences miRBase ver.16 array chips (performed in triplicate). The exoRNAs were hybridized across 1,214 different microRNAs on the probe set.
Of the 1,214 microRNAs analyzed, 79 microRNAs showed a signal intensity >32. Within this set of 79, there was a clear colon versus normal “signature set” of 40 microRNAs that could discriminate normal from colon cancer serum samples with a p-value < 0.01. The identities of the microRNAs found in this study have been masked while further investigation continues.