Exo-Check™ Exosome Antibody Array
- Sensitive—requires as little as 50 µg of exosomal protein for detection
- Convenient—includes eight antibodies for known exosome markers
- Flexible—compatible with most exosome isolation methods, include the ExoQuick® family of reagents and ultracentrifugation
- Complete—includes a control for cellular contamination, a background control (blank spot), and a labeled positive control for HRP detection
- Semi-quantitative—can be used to evaluate relative abundance of certain exosome markers from a given set of samples
Products
| Catalog Number | Description | Size | Price | Quantity | Add to Cart | |||
|---|---|---|---|---|---|---|---|---|
| EXORAY200B-4 | Exo-Check Exosome Antibody Array | 4 Arrays | $572 |
|
||||
| EXORAY210B-8 | Exo-Check Exosome Antibody Array | 8 Arrays | $1058 |
|
||||
Overview
Overview
Streamline your exosome detection with Exo-Check Arrays
For the most efficient detection of exosomes, turn to our semi-quantitative Exo-Check Exosome Antibody Arrays. Each array has 12 pre-printed spots and features 8 antibodies for known exosome markers (CD63, CD81, ALIX, FLOT1, ICAM1, EpCam, ANXA5 and TSG101), a GM130 cis-Golgi marker to monitor any cellular contamination in your exosome isolations, a labeled positive control for HRP detection, and a blank spot as a background control. The kits come complete with a secondary detection mixture conjugated to HRP.
- Sensitive—requires as little as 50 µg of exosomal protein for detection
- Convenient—includes eight antibodies for known exosome markers
- Flexible—compatible with most exosome isolation methods, include the ExoQuick® family of reagents and ultracentrifugation
- Complete—includes a control for cellular contamination, a background control (blank spot), and a labeled positive control for HRP detection
- Semi-quantitative—can be used to evaluate relative abundance of certain exosome markers from a given set of samples
| 1 | 2 | 3 | 4 | 5 | 6 | |
|---|---|---|---|---|---|---|
| A | CD63 | EpCam | ANXA5 | TSG101 | Blank | Positive control |
| B | Positive control | GM130 | FLOT-1 | ICAM | ALIX | CD81 |
| Spot position* | ID | Notes | Link to ExoCarta entry for the human protein |
|---|---|---|---|
| A1 | CD63 | Tetraspanin | ExoCarta_967 |
| A2 | EpCam | Epithelial cell adhesion molecule; often found in cancer-derived exosomes | ExoCarta_4072 |
| A3 | ANXA5 | Annexin A5 | ExoCarta_308 |
| A4 | TSG101 | Tumor susceptibility gene 101 | ExoCarta_7251 |
| A5 | Blank | Background control | NA |
| A6 | Positive control | Positive control for HRP detection (derived from human serum exosomes) | NA |
| B1 | Positive control | Labeled positive control for HRP detection | NA |
| B2 | GM130 | Cis-golgi matrix protein—control for cellular contamination in exosome preparation | NA |
| B3 | FLOT1 | Flotillin-1 | ExoCarta_10211 |
| B4 | ICAM1 | Intercellular adhesion molecule 1 | ExoCarta_3383 |
| B5 | ALIX | Programmed cell death 6 interacting protein (PDCD6IP) | ExoCarta_10015 |
| B6 | CD81 | Tetraspanin | ExoCarta_93185 |
| *To correctly orient the array, place the notched corner in the upper left-hand position |
References
How It Works
Supporting Data
Supporting Data
Streamlined, semi-quantitative exosome detection
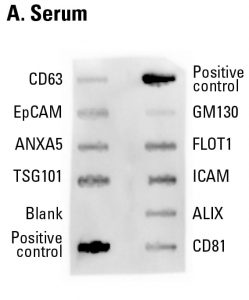
Sample data showing serum-derived exosome detection with an Exo-Check Exosome Antibody Array. The array was exposed to 50 µg of exosome proteins isolated from pooled normal human serum using ExoQuick®. The positive control band should provide strong signals to indicate that all of the detection reagents are working properly. The various exosome antibody spots will provide varying levels of signal, depending upon the source of the isolated exosomes. The blank band serves as a background control and should not have any signal. The GM130 control will only show signal above background if there is cellular contamination in your exosome preparations.
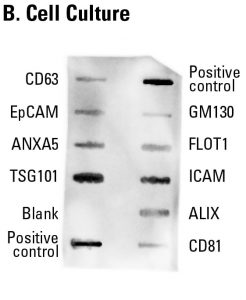
Sample data showing cell culture-derived exosome detection with an Exo-Check Exosome Antibody Array. The array was exposed to 50 µg of exosome proteins isolated from HEK293T cells cultured in SBI’s ExoFBS exosome-depleted media using ExoQuick-TC®. The positive control spots should provide strong signals to indicate that all of the detection reagents are working properly. The various exosome antibody spots will provide varying levels of signal, depending upon the source of the isolated exosomes. The blank band serves as a background control and should not have any signal. The GM130 control will only show signal above background if there is cellular contamination in your exosome preparations.
FAQs
Documentation
Citations
Related Products
Products
| Catalog Number | Description | Size | Price | Quantity | Add to Cart | |||
|---|---|---|---|---|---|---|---|---|
| EXORAY200B-4 | Exo-Check Exosome Antibody Array | 4 Arrays | $572 |
|
||||
| EXORAY210B-8 | Exo-Check Exosome Antibody Array | 8 Arrays | $1058 |
|
||||
Overview
Overview
Streamline your exosome detection with Exo-Check Arrays
For the most efficient detection of exosomes, turn to our semi-quantitative Exo-Check Exosome Antibody Arrays. Each array has 12 pre-printed spots and features 8 antibodies for known exosome markers (CD63, CD81, ALIX, FLOT1, ICAM1, EpCam, ANXA5 and TSG101), a GM130 cis-Golgi marker to monitor any cellular contamination in your exosome isolations, a labeled positive control for HRP detection, and a blank spot as a background control. The kits come complete with a secondary detection mixture conjugated to HRP.
- Sensitive—requires as little as 50 µg of exosomal protein for detection
- Convenient—includes eight antibodies for known exosome markers
- Flexible—compatible with most exosome isolation methods, include the ExoQuick® family of reagents and ultracentrifugation
- Complete—includes a control for cellular contamination, a background control (blank spot), and a labeled positive control for HRP detection
- Semi-quantitative—can be used to evaluate relative abundance of certain exosome markers from a given set of samples
| 1 | 2 | 3 | 4 | 5 | 6 | |
|---|---|---|---|---|---|---|
| A | CD63 | EpCam | ANXA5 | TSG101 | Blank | Positive control |
| B | Positive control | GM130 | FLOT-1 | ICAM | ALIX | CD81 |
| Spot position* | ID | Notes | Link to ExoCarta entry for the human protein |
|---|---|---|---|
| A1 | CD63 | Tetraspanin | ExoCarta_967 |
| A2 | EpCam | Epithelial cell adhesion molecule; often found in cancer-derived exosomes | ExoCarta_4072 |
| A3 | ANXA5 | Annexin A5 | ExoCarta_308 |
| A4 | TSG101 | Tumor susceptibility gene 101 | ExoCarta_7251 |
| A5 | Blank | Background control | NA |
| A6 | Positive control | Positive control for HRP detection (derived from human serum exosomes) | NA |
| B1 | Positive control | Labeled positive control for HRP detection | NA |
| B2 | GM130 | Cis-golgi matrix protein—control for cellular contamination in exosome preparation | NA |
| B3 | FLOT1 | Flotillin-1 | ExoCarta_10211 |
| B4 | ICAM1 | Intercellular adhesion molecule 1 | ExoCarta_3383 |
| B5 | ALIX | Programmed cell death 6 interacting protein (PDCD6IP) | ExoCarta_10015 |
| B6 | CD81 | Tetraspanin | ExoCarta_93185 |
| *To correctly orient the array, place the notched corner in the upper left-hand position |
References
How It Works
Supporting Data
Supporting Data
Streamlined, semi-quantitative exosome detection
Sample data showing serum-derived exosome detection with an Exo-Check Exosome Antibody Array. The array was exposed to 50 µg of exosome proteins isolated from pooled normal human serum using ExoQuick®. The positive control band should provide strong signals to indicate that all of the detection reagents are working properly. The various exosome antibody spots will provide varying levels of signal, depending upon the source of the isolated exosomes. The blank band serves as a background control and should not have any signal. The GM130 control will only show signal above background if there is cellular contamination in your exosome preparations.
Sample data showing cell culture-derived exosome detection with an Exo-Check Exosome Antibody Array. The array was exposed to 50 µg of exosome proteins isolated from HEK293T cells cultured in SBI’s ExoFBS exosome-depleted media using ExoQuick-TC®. The positive control spots should provide strong signals to indicate that all of the detection reagents are working properly. The various exosome antibody spots will provide varying levels of signal, depending upon the source of the isolated exosomes. The blank band serves as a background control and should not have any signal. The GM130 control will only show signal above background if there is cellular contamination in your exosome preparations.