XMIR Texas Red-labeled Positive Control RNA Oligo
Products
| Catalog Number | Description | Size | Price | Quantity | Add to Cart | |||
|---|---|---|---|---|---|---|---|---|
| XMIR-POS | XMIR Texas Red-labeled positive control RNA oligo with Xmotif | 10 Reactions | $562 |
|
||||
Overview
Overview
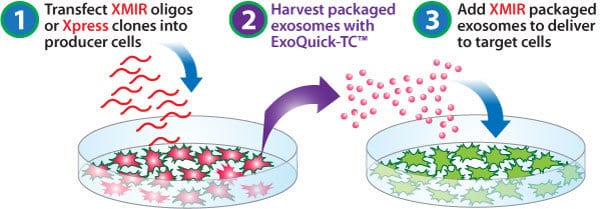
Enhancing your XMIR, AXMIR, and XMIRXpress workflows
Use SBI’s ready-to-transfect XMIR Texas Red-labeled Positive Control Oligo with XMotif as an easy-to-use positive control for your XMIR, AXMIR, and XMIRXpress studies. Simply transfect exosome-producing cells with the XMIR Positive Control Oligo, wait for the oligo to be packaged into exosomes, harvest the exosomes, and incubate with recipient cells. The XMIR Positive Control Oligo will be delivered to the recipient cells, causing them to fluoresce red (use a typical RFP filter set).
References
How It Works
How It Works
Using the XMIR Texas Red-labeled Positive Control Oligo
The XMIR Texas Red-labeled Positive Control Oligo is an RNA oligo containing the fluorescent dye Texas Red conjugated to the XMotif—SBI’s optimized RNA sequence tag that targets a small RNA for exosome packaging. The XMIR Positive Control Oligo is used in the same way as any of our XMIR oligos—simply transfect your exosome-producing cells with the XMIR Positive Control Oligo, wait for the oligo to get packaged into exosomes, harvest exosomes, and add to recipient cells. Your recipient cells will fluoresce red upon successful delivery of the XMIR Positive Control Oligo (works with a typical RFP filter set).
Supporting Data
Supporting Data
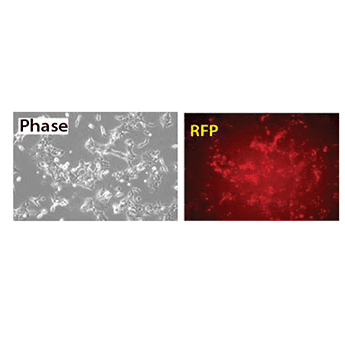
The XMIR Texas Red-labeled Positive Control Oligo in action
Figure 1. The XMIR Texas Red-labeled Positive Control Oligo is efficiently delivered to recipient cells by exosomes. METHODS: HEK-293 cells were transfected with the XMIR Texas Red-labeled Positive Control Oligo. Twenty-four hours later, exosomes were isolated using ExoQuick-TC® and added to naïve HEK293 target cells. After 4-hours, cells were imaged on a Leica DMI300B fluorescence microscope using a standard RFP filter set to visualize the Texas-Red signal.
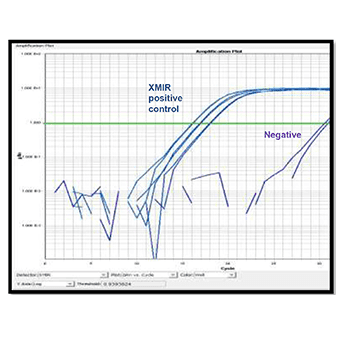
Figure 2. The abundance of the XMIR Texas Red-labeled Positive Control Oligo in recipient cells can be measured using qPCR. METHODS: The recipient HEK-293 cells from Figure 1 were lysed and total cellular RNA extracted in preparation for qPCR. cDNA synthesis and qPCR analysis were performed using using SBI’s QuantiMir kit (Cat.# RA420A-1) and the qPCR assay run on an Applied Biosystems 7900HT qPCR machine using the spike-in forward primer to detect abundance of the positive control oligo. Abundance was normalized to endogenous reference miR-16 levels and calculated using the ΔΔCt method, and clearly show good transfer of the XMIR Positive Control Oligo to the recipient cells.
PRODUCT LIST AND SEARCH BAR:
FAQs
Documentation
Citations
Related Products
Products
| Catalog Number | Description | Size | Price | Quantity | Add to Cart | |||
|---|---|---|---|---|---|---|---|---|
| XMIR-POS | XMIR Texas Red-labeled positive control RNA oligo with Xmotif | 10 Reactions | $562 |
|
||||
Overview
Overview
Enhancing your XMIR, AXMIR, and XMIRXpress workflows
Use SBI’s ready-to-transfect XMIR Texas Red-labeled Positive Control Oligo with XMotif as an easy-to-use positive control for your XMIR, AXMIR, and XMIRXpress studies. Simply transfect exosome-producing cells with the XMIR Positive Control Oligo, wait for the oligo to be packaged into exosomes, harvest the exosomes, and incubate with recipient cells. The XMIR Positive Control Oligo will be delivered to the recipient cells, causing them to fluoresce red (use a typical RFP filter set).
References
How It Works
How It Works
Using the XMIR Texas Red-labeled Positive Control Oligo
The XMIR Texas Red-labeled Positive Control Oligo is an RNA oligo containing the fluorescent dye Texas Red conjugated to the XMotif—SBI’s optimized RNA sequence tag that targets a small RNA for exosome packaging. The XMIR Positive Control Oligo is used in the same way as any of our XMIR oligos—simply transfect your exosome-producing cells with the XMIR Positive Control Oligo, wait for the oligo to get packaged into exosomes, harvest exosomes, and add to recipient cells. Your recipient cells will fluoresce red upon successful delivery of the XMIR Positive Control Oligo (works with a typical RFP filter set).
Supporting Data
Supporting Data
The XMIR Texas Red-labeled Positive Control Oligo in action
Figure 1. The XMIR Texas Red-labeled Positive Control Oligo is efficiently delivered to recipient cells by exosomes. METHODS: HEK-293 cells were transfected with the XMIR Texas Red-labeled Positive Control Oligo. Twenty-four hours later, exosomes were isolated using ExoQuick-TC® and added to naïve HEK293 target cells. After 4-hours, cells were imaged on a Leica DMI300B fluorescence microscope using a standard RFP filter set to visualize the Texas-Red signal.
Figure 2. The abundance of the XMIR Texas Red-labeled Positive Control Oligo in recipient cells can be measured using qPCR. METHODS: The recipient HEK-293 cells from Figure 1 were lysed and total cellular RNA extracted in preparation for qPCR. cDNA synthesis and qPCR analysis were performed using using SBI’s QuantiMir kit (Cat.# RA420A-1) and the qPCR assay run on an Applied Biosystems 7900HT qPCR machine using the spike-in forward primer to detect abundance of the positive control oligo. Abundance was normalized to endogenous reference miR-16 levels and calculated using the ΔΔCt method, and clearly show good transfer of the XMIR Positive Control Oligo to the recipient cells.
PRODUCT LIST AND SEARCH BAR: