LncRNA Profiler qPCR Array Kit (Human)
Products
Overview
Overview
Learn more about lncRNAs
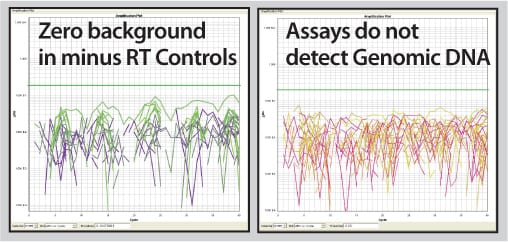
Profile 90 human lncRNAs with this kit that includes reagents for converting lncRNAs to cDNAs and a primer set for qPCR assays in a 96-well format. Get robust cDNA yields and clean, sensitive qPCR quantitation with undetectable background.
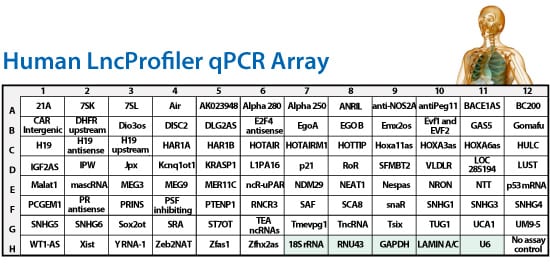 The kit includes five positive housekeeping reference controls and one negative control. In addition, the kit supports subcellular localization studies through RNA reference controls for the nucleolus, nucleus, and cytoplasm:
The kit includes five positive housekeeping reference controls and one negative control. In addition, the kit supports subcellular localization studies through RNA reference controls for the nucleolus, nucleus, and cytoplasm:
- Nucleolus—snoRNA RNU43 (and some 18S rRNA)
- Nucleus—small Nuclear splicing snRNA U6B
- Cytoplasm—GAPDH, Lamin A/C (Human Array) and Beta Actin (Mouse Array) as well as 18S rRNA
References
How It Works
How It Works
Using the LncRNA Profiler qPCR Array Kit
First, get your lncRNA ready for qPCR by converting to cDNA
While some lncRNAs have endogenouse polyA tails, other lncRNAs do not. To enhance qPCR assay performance, the cDNA synthesis workflow includes steps and reagents to polyadenylate all lncRNAs before cDNA conversion using the tagged oligo dT adaptor and random primers. This combined RNA tailing and oligo dT plus random primers boosts cDNA yield significantly and enables strand-specific lncRNA qPCR profiling.
- Tag all lncRNAs with a poly-A tail
- Anneal an oligo-dT adaptor
- Reverse transcribe to create first-strand cDNA
The result is a cDNA pool of anchor-tailed lncRNAs that are ready for qPCR.
Then conduct qPCR assays
Get additional information about the array including lncRNA sequences, LncRNA database links, and qPCR analysis templates in the Excel document, “Assay List and Free Analysis Software for Human LncProfiler qPCR Array ”
Supporting Data
Supporting Data
Example LncRNA Profiler qPCR Array Kit (Human) Data
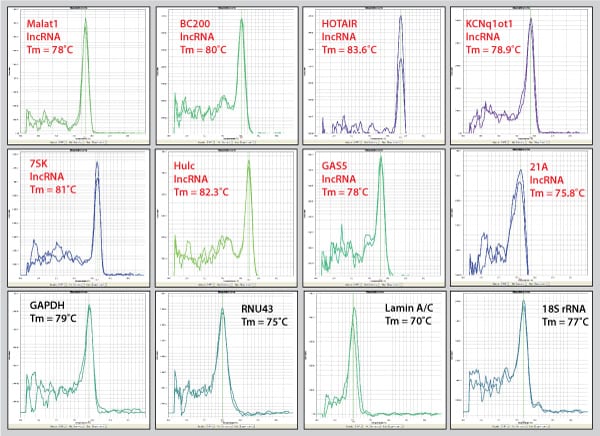
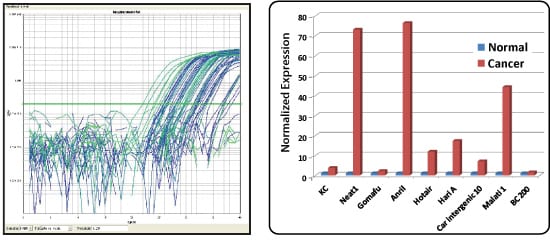
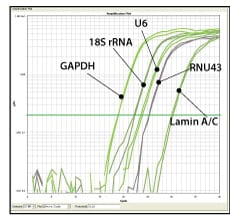
Figure 1. Sample dissociation analysis curves.
Figure 2. Compare lncRNA expression in normal human fibroblasts and fibrosarcoma cells. LncRNAs known to be upregulated in cancer, such as Neat1, Anril, Hotair and Malat1, were all detected at highly elevated levels in the fibrosarcoma cells (HT1080) when compared to the normal fibroblast cells (HFF). The data are plotted as normalized expression levels.
FAQs
Documentation
Citations
Related Products
Products
Overview
Overview
Learn more about lncRNAs
Profile 90 human lncRNAs with this kit that includes reagents for converting lncRNAs to cDNAs and a primer set for qPCR assays in a 96-well format. Get robust cDNA yields and clean, sensitive qPCR quantitation with undetectable background.

 The kit includes five positive housekeeping reference controls and one negative control. In addition, the kit supports subcellular localization studies through RNA reference controls for the nucleolus, nucleus, and cytoplasm:
The kit includes five positive housekeeping reference controls and one negative control. In addition, the kit supports subcellular localization studies through RNA reference controls for the nucleolus, nucleus, and cytoplasm:
- Nucleolus—snoRNA RNU43 (and some 18S rRNA)
- Nucleus—small Nuclear splicing snRNA U6B
- Cytoplasm—GAPDH, Lamin A/C (Human Array) and Beta Actin (Mouse Array) as well as 18S rRNA

References
How It Works
How It Works
Using the LncRNA Profiler qPCR Array Kit
First, get your lncRNA ready for qPCR by converting to cDNA
While some lncRNAs have endogenouse polyA tails, other lncRNAs do not. To enhance qPCR assay performance, the cDNA synthesis workflow includes steps and reagents to polyadenylate all lncRNAs before cDNA conversion using the tagged oligo dT adaptor and random primers. This combined RNA tailing and oligo dT plus random primers boosts cDNA yield significantly and enables strand-specific lncRNA qPCR profiling.
- Tag all lncRNAs with a poly-A tail
- Anneal an oligo-dT adaptor
- Reverse transcribe to create first-strand cDNA
The result is a cDNA pool of anchor-tailed lncRNAs that are ready for qPCR.
Then conduct qPCR assays
Get additional information about the array including lncRNA sequences, LncRNA database links, and qPCR analysis templates in the Excel document, “Assay List and Free Analysis Software for Human LncProfiler qPCR Array ”
Supporting Data
Supporting Data
Example LncRNA Profiler qPCR Array Kit (Human) Data
Figure 1. Sample dissociation analysis curves.
Figure 2. Compare lncRNA expression in normal human fibroblasts and fibrosarcoma cells. LncRNAs known to be upregulated in cancer, such as Neat1, Anril, Hotair and Malat1, were all detected at highly elevated levels in the fibrosarcoma cells (HT1080) when compared to the normal fibroblast cells (HFF). The data are plotted as normalized expression levels.