ExoELISA-ULTRA Complete Kit (CD63 Detection)
- Sensitive—detect as little as 1 µg protein equivalent
- Fast—complete in less than 4-hours—no more overnight incubation
- Flexible—compatible with all major exosome isolation methods (e.g. ExoQuick®, ultracentrifugation, ultrafiltration, and immunoaffinity capture) from human samples
- Quantitative—calibrated internal standards enable quantitation of exosomes carrying CD63
- Sample-saving—requires significantly less sample than our standard ExoELISA Kit, leaving more for other downstream applications
Products
| Catalog Number | Description | Size | Price | Quantity | Add to Cart | |||
|---|---|---|---|---|---|---|---|---|
| EXEL-ULTRA-CD63-1 | ExoELISA-ULTRA Complete Kit (CD63 detection) | 96 Reactions | $937 |
|
||||
Overview
Overview
Delivering ELISA-based exosome quantitation ULTRA fast
Improving on our popular ExoELISA™ Kits, the ExoELISA-ULTRA CD63 Kit increases the sensitivity of exosome detection—as low as 1 µg protein equivalent—while shortening the total assay time to only 4 hours.
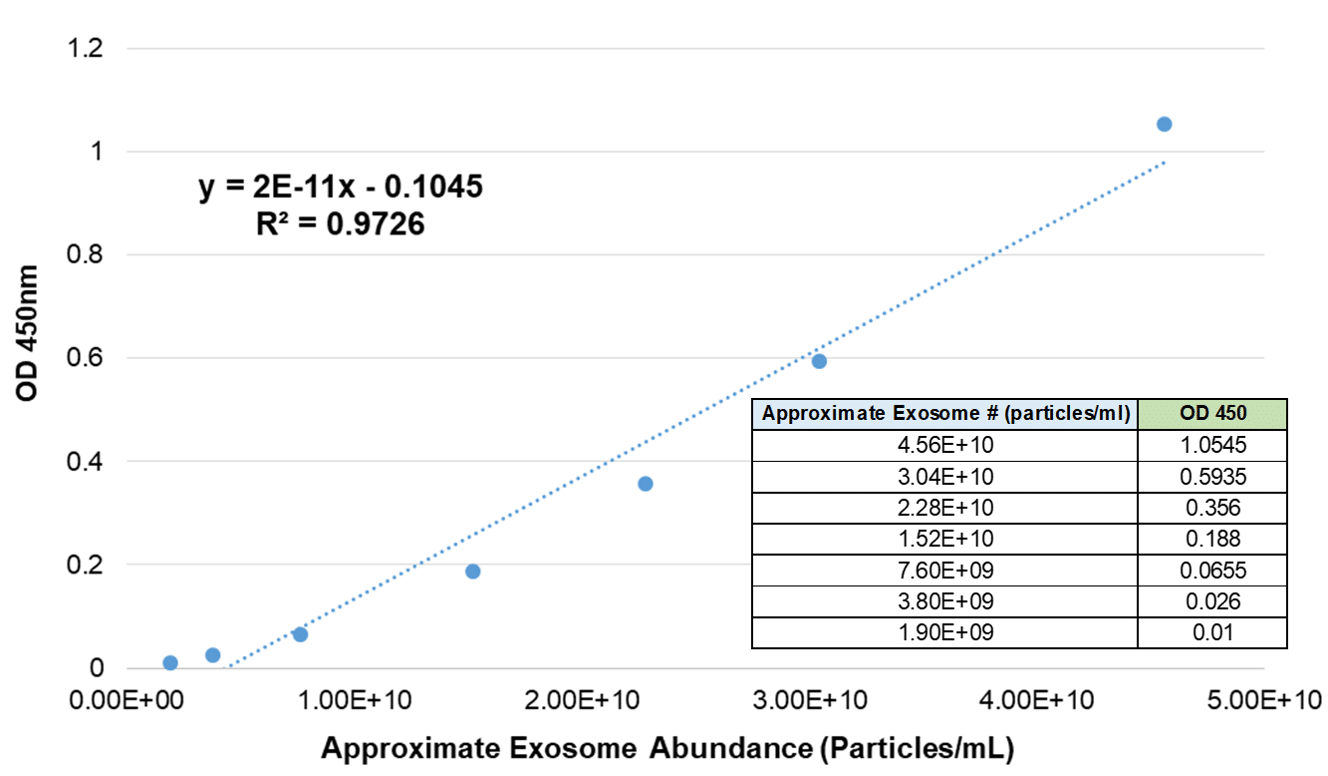
Currently configured for detection of CD63, a widely recognized and popular exosomal marker1, ExoELISA-ULTRA CD63 is based on an ultra-sensitive, direct capture, colorimetric ELISA assay that is compatible with nearly all biofluids. The ExoELISA-ULTRA CD63 Kit comes with an internal standard calibrated to exosomes from a range of biofluids. Calibration is achieved by NanoSight analysis and enables quantitation of exosomes carrying CD63 in your target samples. One ExoELISA-ULTRA CD63 Kit contains all of the necessary reagents (including assay plate) to perform up to 96 reactions.
- Sensitive—detect as little as 1 µg protein equivalent
- Fast—complete in less than 4-hours—no more overnight incubation
- Flexible—compatible with all major exosome isolation methods (e.g. ExoQuick®, ultracentrifugation, ultrafiltration, and immunoaffinity capture) from human samples
- Quantitative—calibrated internal standards enable quantitation of exosomes carrying CD63
- Sample-saving—requires significantly less sample than our standard ExoELISA Kit, leaving more for other downstream applications
| ExoELISA-ULTRA Complete Kits | EXOCET | FluoroCet | |
|---|---|---|---|
| Use | For fast and sensitive antibody-based quantitation of exosomes | For fast quantitation of extracellular vesicles with moderate sample input requirements | For the most sensitive quantitation of extracellular vesicles with very low sample input requirements |
| Detection method | Antibody | Enzymatic | Enzymatic |
| Quantitation chemistry | Enzymatic (HRP) | Colorimetric | Fluorescent |
| Total protocol time | 4 hours (no overnight incubation) | 20 min | 60 min |
| Input sample amount (protein equivalent) | 1 – 200 µg | 50 µg | <1 µg |
| Learn More | ExoELISA-ULTRA CD63 ExoELISA-ULTRA CD81 ExoELISA-ULTRA CD9 ExoELISA-ULTRA GroEL | EXOCET | FluoroCet |
- Kowal, J., et al. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci U S A. 2016. February 23. 113(8): E968–E977. PMCID: PMC4776515.
References
How It Works
Supporting Data
FAQs
Documentation
Citations
Related Products
Products
| Catalog Number | Description | Size | Price | Quantity | Add to Cart | |||
|---|---|---|---|---|---|---|---|---|
| EXEL-ULTRA-CD63-1 | ExoELISA-ULTRA Complete Kit (CD63 detection) | 96 Reactions | $937 |
|
||||
Overview
Overview
Delivering ELISA-based exosome quantitation ULTRA fast
Improving on our popular ExoELISA™ Kits, the ExoELISA-ULTRA CD63 Kit increases the sensitivity of exosome detection—as low as 1 µg protein equivalent—while shortening the total assay time to only 4 hours.
Currently configured for detection of CD63, a widely recognized and popular exosomal marker1, ExoELISA-ULTRA CD63 is based on an ultra-sensitive, direct capture, colorimetric ELISA assay that is compatible with nearly all biofluids. The ExoELISA-ULTRA CD63 Kit comes with an internal standard calibrated to exosomes from a range of biofluids. Calibration is achieved by NanoSight analysis and enables quantitation of exosomes carrying CD63 in your target samples. One ExoELISA-ULTRA CD63 Kit contains all of the necessary reagents (including assay plate) to perform up to 96 reactions.
- Sensitive—detect as little as 1 µg protein equivalent
- Fast—complete in less than 4-hours—no more overnight incubation
- Flexible—compatible with all major exosome isolation methods (e.g. ExoQuick®, ultracentrifugation, ultrafiltration, and immunoaffinity capture) from human samples
- Quantitative—calibrated internal standards enable quantitation of exosomes carrying CD63
- Sample-saving—requires significantly less sample than our standard ExoELISA Kit, leaving more for other downstream applications
| ExoELISA-ULTRA Complete Kits | EXOCET | FluoroCet | |
|---|---|---|---|
| Use | For fast and sensitive antibody-based quantitation of exosomes | For fast quantitation of extracellular vesicles with moderate sample input requirements | For the most sensitive quantitation of extracellular vesicles with very low sample input requirements |
| Detection method | Antibody | Enzymatic | Enzymatic |
| Quantitation chemistry | Enzymatic (HRP) | Colorimetric | Fluorescent |
| Total protocol time | 4 hours (no overnight incubation) | 20 min | 60 min |
| Input sample amount (protein equivalent) | 1 – 200 µg | 50 µg | <1 µg |
| Learn More | ExoELISA-ULTRA CD63 ExoELISA-ULTRA CD81 ExoELISA-ULTRA CD9 ExoELISA-ULTRA GroEL | EXOCET | FluoroCet |
- Kowal, J., et al. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci U S A. 2016. February 23. 113(8): E968–E977. PMCID: PMC4776515.