OmniTag System
- Universal Minicircle Donor that work for tagging any gene
- No need to prepare target-specific homology donor constructs
- Foreign-DNA free, ready to use Minicircle Donors
- Simplicity and modularity at high efficiency
Products
| Catalog Number | Description | Size | Price | Quantity | Add to Cart | |||
|---|---|---|---|---|---|---|---|---|
| OT100-F0 | OmniTag F0 Frame Selector Vector with CMV-hspCas | 10 µg | $1054 |
|
||||
| OT101-F1 | OmniTag F1 Frame Selector Vector with CMV-hspCas9 | 10 µg | $1054 |
|
||||
| OT102-F2 | OmniTag F2 Frame Selector Vector with CMV-hspCas9 | 10 µg | $1054 |
|
||||
| OT200-F0 | OmniTag F0 Frame Selector Vector with EF1alpha-hspCas9 | 10 µg | $1054 |
|
||||
| OT201-F1 | OmniTag F1 Frame Selector Vector with EF1alpha-hspCas9 | 10 µg | $1054 |
|
||||
| OT202-F2 | OmniTag F2 Frame Selector Vector with EF1alpha-hspCas9 | 10 µg | $1054 |
|
||||
| OT501MC-1 | copGFP-T2A-Puro-pA OmniTag Minicircle Donor | 10 µg | $1054 |
|
||||
| OT502MC-1 | RFP-T2A-Blast-pA OmniTag Minicircle Donor | 10 µg | $1054 |
|
||||
| OT503MC-1 | 6xHis-T2A-Puro-pA OmniTag Minicircle Donor | 10 µg | $1054 |
|
||||
| OT504MC-1 | 3xFlag-T2A-Puro-pA OmniTag Minicircle Donor | 10 µg | $1054 |
|
||||
* With the help of SBI’s highly affordable Syn2Clone service, you can order any OmniTag Minicircle Donor with your desired tag and selection marker of your choice and have it made On-Demand very swiftly.
** Please inquire about bundle quote at discounted pricing for buying all three Frame Selector Vectors.
Overview
Overview
SBI’s OmniTag System streamlines tagging any endogenous gene at its C-terminus when used with one of the OmniTag F0, F1 or F2 Frame Selector Vectors and OmniTag Minicircle Donor vector of your choice.
- Universal Minicircle Donor that work for tagging any gene
- No need to prepare target-specific homology arms
- Foreign-DNA free, ready to use Minicircle Donors
- Frame selector vectors to specify the insertion reading frame
- CMV or EF1alpha promoter options to express Cas9 from Frame Selector
- Simplicity and modularity at high efficiency - >90% tag-positive cells following antibiotic selection
- Multiplex- sequentially tag multiple genes in same cells with choice of appropriate donors
- Alleviate the requirement for protein-specific antibodies for endogenous gene expression quantification, protein localization or immuno-precipitation studies.
Possible Applications
Tagging endogenous genes at their C- terminus by editing the genomic locus opens up plethora of possible applications and offers advantages to researchers in various areas of research.
- Tagging with Luciferase (T2A-Gaussia Luciferase, NanoLuc) for expression measurement
- Tagging with various fluorescent proteins (TagGFP2, TagBFP, TagRFP and T2A-TurboGFP-PEST) for cellular localization and FACS
- Tagging with small epitope tags (HA, Myc, Strep tag II, Streptavidin-binding peptide, AviTag, SpyTag, SunTag) for sensitive detection or affinity purification approaches
- Tagging with a controllable destabilization tag (ProtoTuner DD) to achieve switchable gene expression
- Tagging with split tags (split-TEV protease) to assess protein–protein interaction
Why choose OmniTag?
Tagging endogenous genes at their C- terminus by editing the genomic locus meets with the challenge of removing stop codon and in frame editing so that the tag is fused and translated along with the protein of interest. SBI’s OmniTag System addresses these challenges to simplify C-Terminal tagging of any endogenous gene.
OmniTag Minicircle Donor are minicircle DNAs that will be fully integrated in the genomic locus and offer important features such as
- Foreign DNA-free- no bacterial plasmid DNA sequences so low immune response
- More efficient transfections due to small size- more copies per ug of DNA
References
dJonathan L. Schmid-Burgk, Klara Höning, Thomas S. Ebert & Veit Hornung. CRISPaint allows modular base-specific gene tagging using a ligase-4 dependent mechanism. Nat Commun. 2016 Jul 28;7:12338. doi:10.1038/ncomms12338. PMCID: PMC4974478
References
How It Works
How It Works
OmniTag System is composed of 3 key components.
- An all-in-one Cas9/gRNA vector, similar to SBI’s CAS7XXX-1, to introduce a double stranded break at the genomic locus just upstream of stop codon of the gene of interest.
- One of the three OmniTag Frame Selector Vectors that are also an all-in-one vector that contains Cas9 and specific gRNAs against frame selector linker in the OmniTag Donor at three possible positions (F0, F1, F2) to achieve in-frame fusion of the tag.
- An OmniTag Donor that is a minicircle DNA containing a frame selector linker, desired tag sequence and a selection marker separated by T2A which gets fully incorporated at the genomic locus by NHEJ after linearization at the frame selector linker.
When these three components are co-transfected into target cells they work in a concerted fashion to achieve C-terminal tagging at genomic locus via a ligase-4-dependent non-homologous end joining (NHEJ).
C-terminal Gene Tagging with OmniTag System
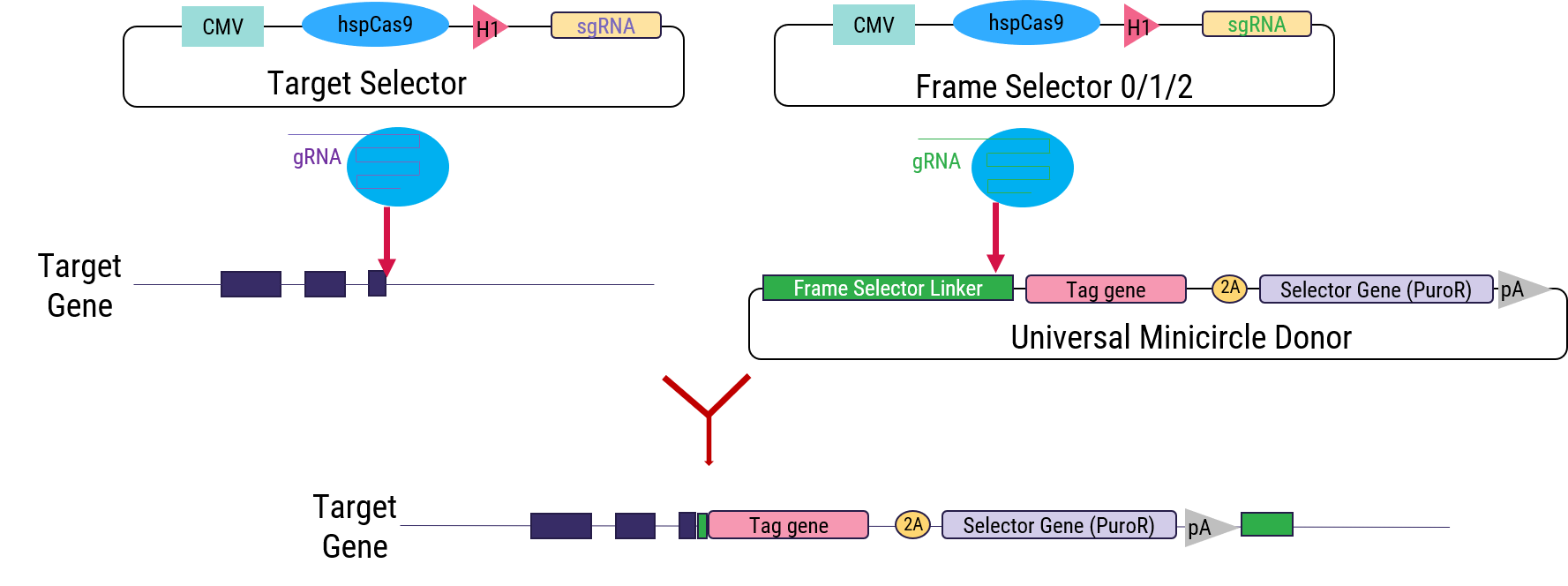
Figure 1. C Terminal tagging an endogenous gene at its genomic locus using OmniTag System
The workflow at-a-glance
CHOOSE: target gRNA and OmniTag Frame Selector vector
To simplify the application of C-terminal tagging using OmniTag System, a list of pre-selected, sequence-optimized target sites close to translation termination codons of nearly all human and mouse protein coding genes in conjunction with the required frame selector can be found here.
CONSTRUCT: Clone target gRNA into all-in-one Cas9 vector such as CAS7XXX-1 as Target Selector
CO-TRANSFECT: Introduce Cas9, target gRNA, OmniTag Frame Selector Vector and OmniTag Donor into the target cells using co-transfection
SELECT/SCREEN: Select or screen clones for verification.
VALIDATE: Genotype/Fluorescent microscopy/Western Blot/Sequencing.
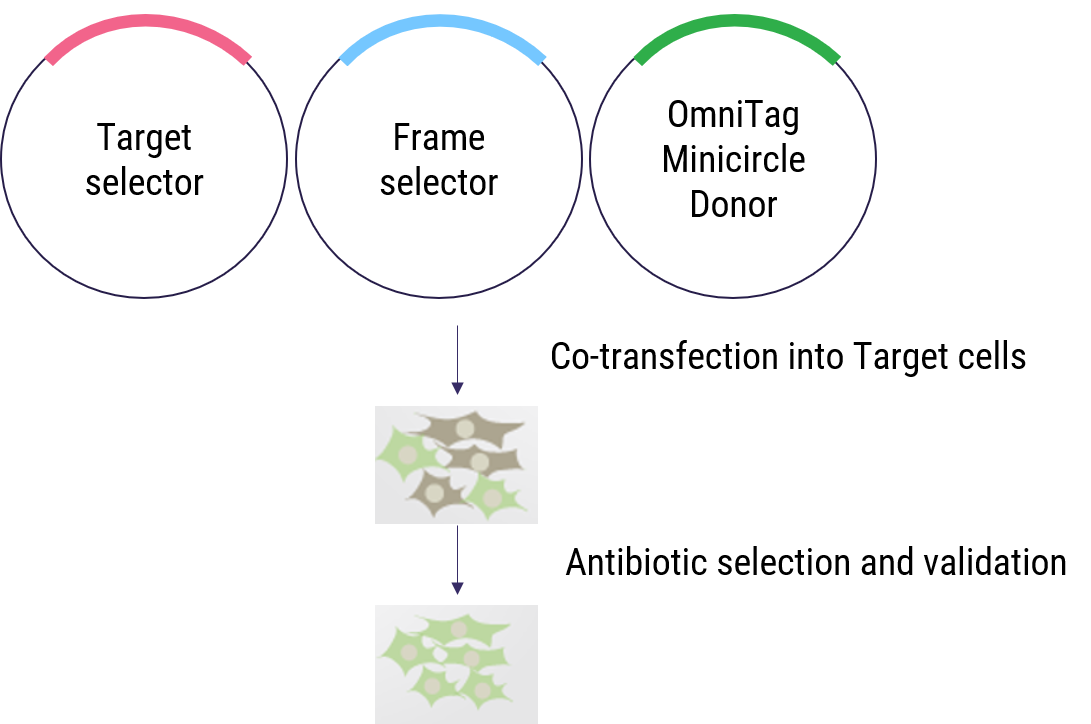
Figure 2. Workflow of OmniTag system for selection-based gene tagging in target cells.
Supporting Data
Tagging of endogenous TUBB with GFP
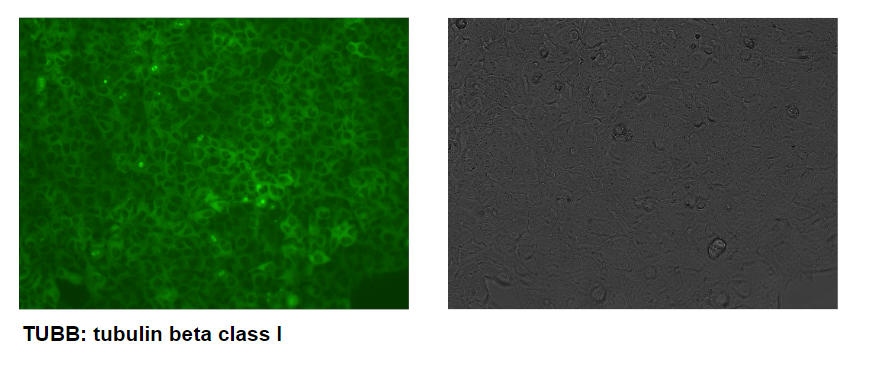
Figure 3. Using OmniTag system to tag endogenous TUBB. HEK293 cells were transfected with Cas9 all-in-one vector containing gRNA targeting TUBB, F1 Frame selector and OmniTag GFP-Puro minicircle donor. 2-4 days after transfection, puromycin at 1ug/ml concentration was applied to cell culture medium. After 4-7 days, puromycin positive clones can be obtained. Fluorescent microscope reveals the GFP tagging of TUBB with corresponding cellular localization.
Confirmation of precise integration of OmniTag Minicircle donor
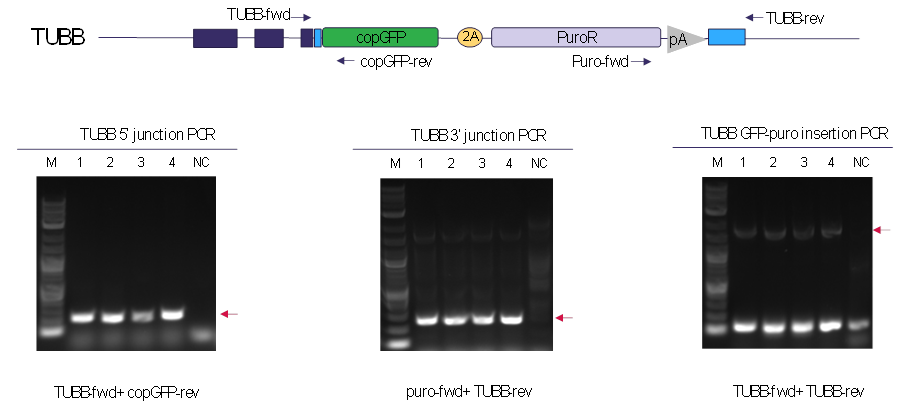
Figure 4. Verification of precise tagging with OmniTag system by junction and insertion PCR. Junction and insertion PCR were performed to confirm the correct integration of OmniTag minicircle donor at the TUBB target site. After puromycin selection, individual clones were expanded, followed by genomic DNA extraction and junction PCR. TUBB-fwd and copGFP-rev primers were used for 5’ junction PCR, puro-fwd and TUBB-rev primers were used for 3’ junction PCR. TUBB-fwd and TUBB-rev primers were used for minicircle donor insertion at TUBB target locus. Gel electrophoresis reveals all the clones contain the expected amplicon, indicating the precise insertion of the OmniTag minicircle donor at the desired TUBB target site.
Dual Tagging of H4C3 and TUBB with different fluorescent tags sequentially
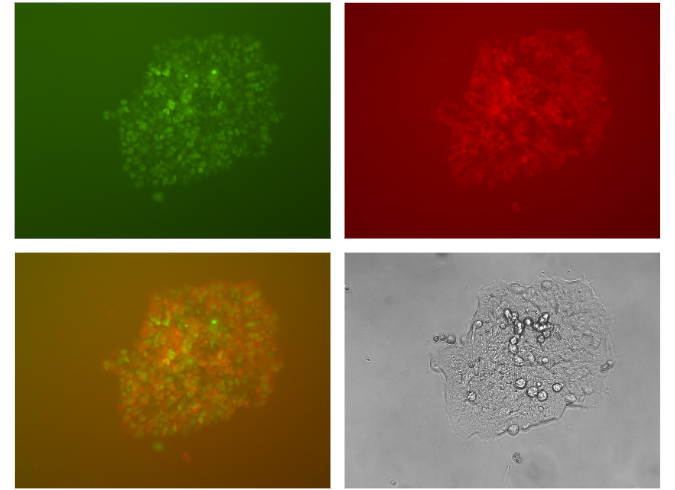
Figure 5. Dual tagging achieved by OmniTag system. HEK293 cells were first transfected with Cas9 all-in-one vector containing sgRNA targeting H4C3, F1 Frame selector and OmniTag GFP-Puro minicircle donor. 2-4 days after transfection, puromycin at 1ug/ml concentration was applied to cell culture medium. After 4-7 days, puromycin positive clones can be obtained. Then HEK293 H4C3-GFP tagging cells were further transfected with Cas9 all-in-one vector containing sgRNA targeting TUBB, F1 Frame selector and OmniTag RFP-Blast minicircle donor. 2-4 days after transfection, blasticidin at 7ug/ml concentration was applied to cell culture medium. After 4-7 days, blasticidin positive clones can be obtained. Fluorescent microscope reveals the RFP tagging of TUBB and GFP tagging of H4C3 with corresponding cellular localization.
FAQs
Documentation
Citations
Related Products
Products
| Catalog Number | Description | Size | Price | Quantity | Add to Cart | |||
|---|---|---|---|---|---|---|---|---|
| OT100-F0 | OmniTag F0 Frame Selector Vector with CMV-hspCas | 10 µg | $1054 |
|
||||
| OT101-F1 | OmniTag F1 Frame Selector Vector with CMV-hspCas9 | 10 µg | $1054 |
|
||||
| OT102-F2 | OmniTag F2 Frame Selector Vector with CMV-hspCas9 | 10 µg | $1054 |
|
||||
| OT200-F0 | OmniTag F0 Frame Selector Vector with EF1alpha-hspCas9 | 10 µg | $1054 |
|
||||
| OT201-F1 | OmniTag F1 Frame Selector Vector with EF1alpha-hspCas9 | 10 µg | $1054 |
|
||||
| OT202-F2 | OmniTag F2 Frame Selector Vector with EF1alpha-hspCas9 | 10 µg | $1054 |
|
||||
| OT501MC-1 | copGFP-T2A-Puro-pA OmniTag Minicircle Donor | 10 µg | $1054 |
|
||||
| OT502MC-1 | RFP-T2A-Blast-pA OmniTag Minicircle Donor | 10 µg | $1054 |
|
||||
| OT503MC-1 | 6xHis-T2A-Puro-pA OmniTag Minicircle Donor | 10 µg | $1054 |
|
||||
| OT504MC-1 | 3xFlag-T2A-Puro-pA OmniTag Minicircle Donor | 10 µg | $1054 |
|
||||
* With the help of SBI’s highly affordable Syn2Clone service, you can order any OmniTag Minicircle Donor with your desired tag and selection marker of your choice and have it made On-Demand very swiftly.
** Please inquire about bundle quote at discounted pricing for buying all three Frame Selector Vectors.
Overview
Overview
SBI’s OmniTag System streamlines tagging any endogenous gene at its C-terminus when used with one of the OmniTag F0, F1 or F2 Frame Selector Vectors and OmniTag Minicircle Donor vector of your choice.
- Universal Minicircle Donor that work for tagging any gene
- No need to prepare target-specific homology arms
- Foreign-DNA free, ready to use Minicircle Donors
- Frame selector vectors to specify the insertion reading frame
- CMV or EF1alpha promoter options to express Cas9 from Frame Selector
- Simplicity and modularity at high efficiency - >90% tag-positive cells following antibiotic selection
- Multiplex- sequentially tag multiple genes in same cells with choice of appropriate donors
- Alleviate the requirement for protein-specific antibodies for endogenous gene expression quantification, protein localization or immuno-precipitation studies.
Possible Applications
Tagging endogenous genes at their C- terminus by editing the genomic locus opens up plethora of possible applications and offers advantages to researchers in various areas of research.
- Tagging with Luciferase (T2A-Gaussia Luciferase, NanoLuc) for expression measurement
- Tagging with various fluorescent proteins (TagGFP2, TagBFP, TagRFP and T2A-TurboGFP-PEST) for cellular localization and FACS
- Tagging with small epitope tags (HA, Myc, Strep tag II, Streptavidin-binding peptide, AviTag, SpyTag, SunTag) for sensitive detection or affinity purification approaches
- Tagging with a controllable destabilization tag (ProtoTuner DD) to achieve switchable gene expression
- Tagging with split tags (split-TEV protease) to assess protein–protein interaction
Why choose OmniTag?
Tagging endogenous genes at their C- terminus by editing the genomic locus meets with the challenge of removing stop codon and in frame editing so that the tag is fused and translated along with the protein of interest. SBI’s OmniTag System addresses these challenges to simplify C-Terminal tagging of any endogenous gene.
OmniTag Minicircle Donor are minicircle DNAs that will be fully integrated in the genomic locus and offer important features such as
- Foreign DNA-free- no bacterial plasmid DNA sequences so low immune response
- More efficient transfections due to small size- more copies per ug of DNA
References
dJonathan L. Schmid-Burgk, Klara Höning, Thomas S. Ebert & Veit Hornung. CRISPaint allows modular base-specific gene tagging using a ligase-4 dependent mechanism. Nat Commun. 2016 Jul 28;7:12338. doi:10.1038/ncomms12338. PMCID: PMC4974478
References
How It Works
How It Works
OmniTag System is composed of 3 key components.
- An all-in-one Cas9/gRNA vector, similar to SBI’s CAS7XXX-1, to introduce a double stranded break at the genomic locus just upstream of stop codon of the gene of interest.
- One of the three OmniTag Frame Selector Vectors that are also an all-in-one vector that contains Cas9 and specific gRNAs against frame selector linker in the OmniTag Donor at three possible positions (F0, F1, F2) to achieve in-frame fusion of the tag.
- An OmniTag Donor that is a minicircle DNA containing a frame selector linker, desired tag sequence and a selection marker separated by T2A which gets fully incorporated at the genomic locus by NHEJ after linearization at the frame selector linker.
When these three components are co-transfected into target cells they work in a concerted fashion to achieve C-terminal tagging at genomic locus via a ligase-4-dependent non-homologous end joining (NHEJ).
C-terminal Gene Tagging with OmniTag System

Figure 1. C Terminal tagging an endogenous gene at its genomic locus using OmniTag System
The workflow at-a-glance
CHOOSE: target gRNA and OmniTag Frame Selector vector
To simplify the application of C-terminal tagging using OmniTag System, a list of pre-selected, sequence-optimized target sites close to translation termination codons of nearly all human and mouse protein coding genes in conjunction with the required frame selector can be found here.
CONSTRUCT: Clone target gRNA into all-in-one Cas9 vector such as CAS7XXX-1 as Target Selector
CO-TRANSFECT: Introduce Cas9, target gRNA, OmniTag Frame Selector Vector and OmniTag Donor into the target cells using co-transfection
SELECT/SCREEN: Select or screen clones for verification.
VALIDATE: Genotype/Fluorescent microscopy/Western Blot/Sequencing.

Figure 2. Workflow of OmniTag system for selection-based gene tagging in target cells.
Supporting Data
Tagging of endogenous TUBB with GFP

Figure 3. Using OmniTag system to tag endogenous TUBB. HEK293 cells were transfected with Cas9 all-in-one vector containing gRNA targeting TUBB, F1 Frame selector and OmniTag GFP-Puro minicircle donor. 2-4 days after transfection, puromycin at 1ug/ml concentration was applied to cell culture medium. After 4-7 days, puromycin positive clones can be obtained. Fluorescent microscope reveals the GFP tagging of TUBB with corresponding cellular localization.
Confirmation of precise integration of OmniTag Minicircle donor

Figure 4. Verification of precise tagging with OmniTag system by junction and insertion PCR. Junction and insertion PCR were performed to confirm the correct integration of OmniTag minicircle donor at the TUBB target site. After puromycin selection, individual clones were expanded, followed by genomic DNA extraction and junction PCR. TUBB-fwd and copGFP-rev primers were used for 5’ junction PCR, puro-fwd and TUBB-rev primers were used for 3’ junction PCR. TUBB-fwd and TUBB-rev primers were used for minicircle donor insertion at TUBB target locus. Gel electrophoresis reveals all the clones contain the expected amplicon, indicating the precise insertion of the OmniTag minicircle donor at the desired TUBB target site.
Dual Tagging of H4C3 and TUBB with different fluorescent tags sequentially

Figure 5. Dual tagging achieved by OmniTag system. HEK293 cells were first transfected with Cas9 all-in-one vector containing sgRNA targeting H4C3, F1 Frame selector and OmniTag GFP-Puro minicircle donor. 2-4 days after transfection, puromycin at 1ug/ml concentration was applied to cell culture medium. After 4-7 days, puromycin positive clones can be obtained. Then HEK293 H4C3-GFP tagging cells were further transfected with Cas9 all-in-one vector containing sgRNA targeting TUBB, F1 Frame selector and OmniTag RFP-Blast minicircle donor. 2-4 days after transfection, blasticidin at 7ug/ml concentration was applied to cell culture medium. After 4-7 days, blasticidin positive clones can be obtained. Fluorescent microscope reveals the RFP tagging of TUBB and GFP tagging of H4C3 with corresponding cellular localization.

