Stem Cell miRNA Profiling Panel
Products
Overview
Overview
Easily monitor miRNAs involved in differentiation
With qPCR primers for a carefully curated set of miRNAs involved in stem cell self-renewal, hematopoiesis, neural development, and tissue patterning, and SBI’s sensitive and reliable RNA-Quant™ cDNA Synthesis Kit, our Stem Cell miRNA Profiling Panel provides a streamlined system for studying differentiation with qPCR. The Stem Cell miRNA Profiling Panel can simultaneously quantitate relative expression level differences between two or more samples for 95 separate markers, with all miRNAS carefully selected from the published literature.
- Simultaneously profile 95 different miRNAs known to be involved in stem cell self-renewal, hematopoiesis, neural development, and tissue patterning
- Identify miRNA biomarkers and expression pattern signatures
- Rapidly tag and convert all small RNAs into detectable cDNA for qPCR
- Measure as little as picogram amounts of starting total RNA
- Conduct high-throughput screens of cell lines and tissues
About the included RNA-Quant cDNA Synthesis Kit
With a single cDNA synthesis reaction, you can efficiently prepare all of the RNAs in your sample for real-time qPCR using the RNA-Quant cDNA Synthesis Kit. Unlike other cDNA synthesis kits which can only prepare one or two classes of RNAs for qPCR, the RNA-Quant Kit enables qPCR measurement of many classes of RNA from a single cDNA synthesis reaction. The kit includes three endogenous RNA reference qPCR assays as for human and mouse samples—miR-16, Y RNA lncRNA, and GAPDH mRNA.
An all-in-one cDNA synthesis kit
- Tail and tag all RNAs for cDNA synthesis
- Obtain full RNA coverage for any purpose
- Profile any RNA—mRNA, miRNA, lncRNA, sn/snoRNA, rRNA
References
How It Works
How It Works
Using the OncoMir qPCR Array
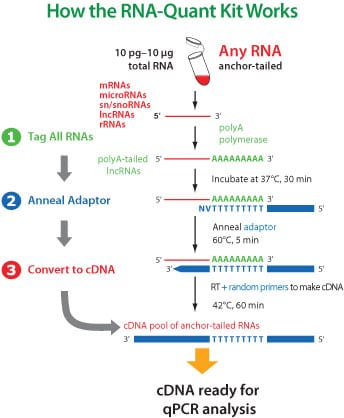
First, prepare RNA for qPCR using the RNA-Quant cDNA Synthesis Kit
The poly-A tailing and tagging method in the RNA-Quant System generates cDNA that is highly unique and, when used with the 3’ Universal Reverse Primer, does not produce any signal in the absence of reverse transcriptase or lead to the detection of any residual genomic DNA. The result is zero background for RNA profiling with complete confidence.
- Tag all small RNAs with a poly-A tail
- Anneal an oligo-dT adaptor
- Reverse transcribe to create first-strand cDNA
Your final product will be a cDNA pool of anchor-tailed miRNAs that are ready for qPCR.
Then conduct qPCR assays using the 96-well primer array
Stem cell signature set: A1 – C5
Hematopoietic set: C6 – F8
Neuronal set: F9 – H5
Organ set: H6 – H11
Control: H12
Get additional information about the array including miRNA sequences, miRbase accession numbers, and qPCR analysis templates in the Excel document, “Free Analysis Software for Stem Cell qPCR Array”
Supporting Data
Supporting Data
Example Stem Cell miRNA Profiling Panel Data
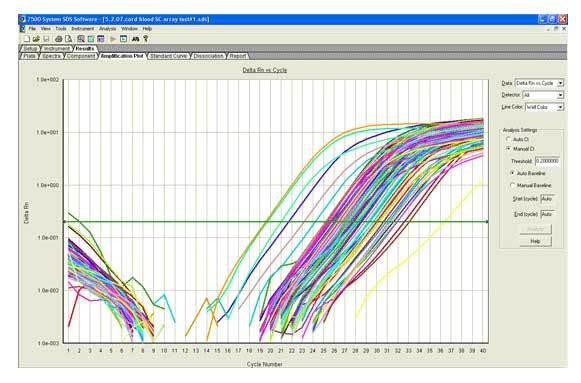
The Stem Cells microRNA qPCR Panel was used to quantitate microRNAs in Human Cord Blood CD34+ Stem Cells.
Figure 1. Real-time amplification plot.
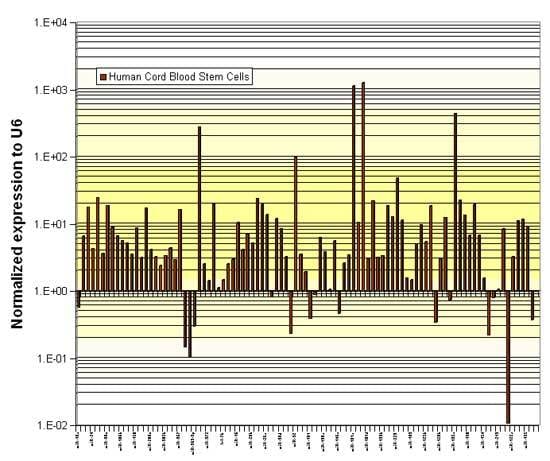
Figure 2. Relative abundance of miRNAs in human cord blood CD34+ stem cells (levels normalized to U6).
FAQs
Documentation
Citations
Related Products
Products
Overview
Overview
Easily monitor miRNAs involved in differentiation
With qPCR primers for a carefully curated set of miRNAs involved in stem cell self-renewal, hematopoiesis, neural development, and tissue patterning, and SBI’s sensitive and reliable RNA-Quant™ cDNA Synthesis Kit, our Stem Cell miRNA Profiling Panel provides a streamlined system for studying differentiation with qPCR. The Stem Cell miRNA Profiling Panel can simultaneously quantitate relative expression level differences between two or more samples for 95 separate markers, with all miRNAS carefully selected from the published literature.
- Simultaneously profile 95 different miRNAs known to be involved in stem cell self-renewal, hematopoiesis, neural development, and tissue patterning
- Identify miRNA biomarkers and expression pattern signatures
- Rapidly tag and convert all small RNAs into detectable cDNA for qPCR
- Measure as little as picogram amounts of starting total RNA
- Conduct high-throughput screens of cell lines and tissues
About the included RNA-Quant cDNA Synthesis Kit
With a single cDNA synthesis reaction, you can efficiently prepare all of the RNAs in your sample for real-time qPCR using the RNA-Quant cDNA Synthesis Kit. Unlike other cDNA synthesis kits which can only prepare one or two classes of RNAs for qPCR, the RNA-Quant Kit enables qPCR measurement of many classes of RNA from a single cDNA synthesis reaction. The kit includes three endogenous RNA reference qPCR assays as for human and mouse samples—miR-16, Y RNA lncRNA, and GAPDH mRNA.
An all-in-one cDNA synthesis kit
- Tail and tag all RNAs for cDNA synthesis
- Obtain full RNA coverage for any purpose
- Profile any RNA—mRNA, miRNA, lncRNA, sn/snoRNA, rRNA
References
How It Works
How It Works
Using the OncoMir qPCR Array
First, prepare RNA for qPCR using the RNA-Quant cDNA Synthesis Kit
The poly-A tailing and tagging method in the RNA-Quant System generates cDNA that is highly unique and, when used with the 3’ Universal Reverse Primer, does not produce any signal in the absence of reverse transcriptase or lead to the detection of any residual genomic DNA. The result is zero background for RNA profiling with complete confidence.
- Tag all small RNAs with a poly-A tail
- Anneal an oligo-dT adaptor
- Reverse transcribe to create first-strand cDNA
Your final product will be a cDNA pool of anchor-tailed miRNAs that are ready for qPCR.
Then conduct qPCR assays using the 96-well primer array
Stem cell signature set: A1 – C5
Hematopoietic set: C6 – F8
Neuronal set: F9 – H5
Organ set: H6 – H11
Control: H12
Get additional information about the array including miRNA sequences, miRbase accession numbers, and qPCR analysis templates in the Excel document, “Free Analysis Software for Stem Cell qPCR Array”
Supporting Data
Supporting Data
Example Stem Cell miRNA Profiling Panel Data
The Stem Cells microRNA qPCR Panel was used to quantitate microRNAs in Human Cord Blood CD34+ Stem Cells.
Figure 1. Real-time amplification plot.

Figure 2. Relative abundance of miRNAs in human cord blood CD34+ stem cells (levels normalized to U6).