PiggyBac qPCR Copy Number Kit
Products
| Catalog Number | Description | Size | Price | Quantity | Add to Cart | |||
|---|---|---|---|---|---|---|---|---|
| PBC100A-1 | piggyBac qPCR copy number kit, 20 reactions | 20 Reactions | $460 |
|
||||
Overview
Overview
Confidently count your PiggyBac insertsWhen you need to know how many PiggyBac integration events have happened, turn to SBI’s PiggyBac qPCR Copy Number Kit. This kit uses qPCR to measure the number of PiggyBac inserts relative to a specific genomic locus—the UCR1 element—with the PiggyBac insert copy number calculated using the cycle threshold (Ct) values of the UCR1 signal relative to the PiggyBac insert signal.
The PiggyBac qPCR Copy Number Kit comes with enough reagents—UCR1 primers, PiggyBac primers, and cell lysis buffer—for twenty copy number determinations, and is compatible with all of SBI’s PiggyBac Vectors.
NOTE: Your cells must be passaged at least once before performing this copy number measurement to ensure that residual, non-integrated piggyBac transposon plasmid does not interfere with the qPCR reaction.
References
How It Works
How It Works
Calculate PiggyBac insert copy number
The PiggyBac qPCR Copy Number Kit provides robust PiggyBac insert copy number determination. To calculate the PiggyBac insert copy number from the Ct values:
- Calculate the average Cts for the PiggyBac inserts and for the UCR1 loci (there are two UCR1 elements per genome)
- The copy number is the ΔΔCt/2—the ΔΔCt value must be divided by two to account for the two UCR1 elements:
ΔΔCt = 2-((average PiggyBac insert Ct) – (average UCR1 Ct))
PiggyBac insert copy number = ΔΔCt/2
You can change the number of PiggyBac insertions by adjusting the ratio of PiggyBac Vector to Super PiggyBac Transposase Expression Vector (Cat.# PB210PA-1).
Supporting Data
Supporting Data
Robust PiggyBac insert copy number determination
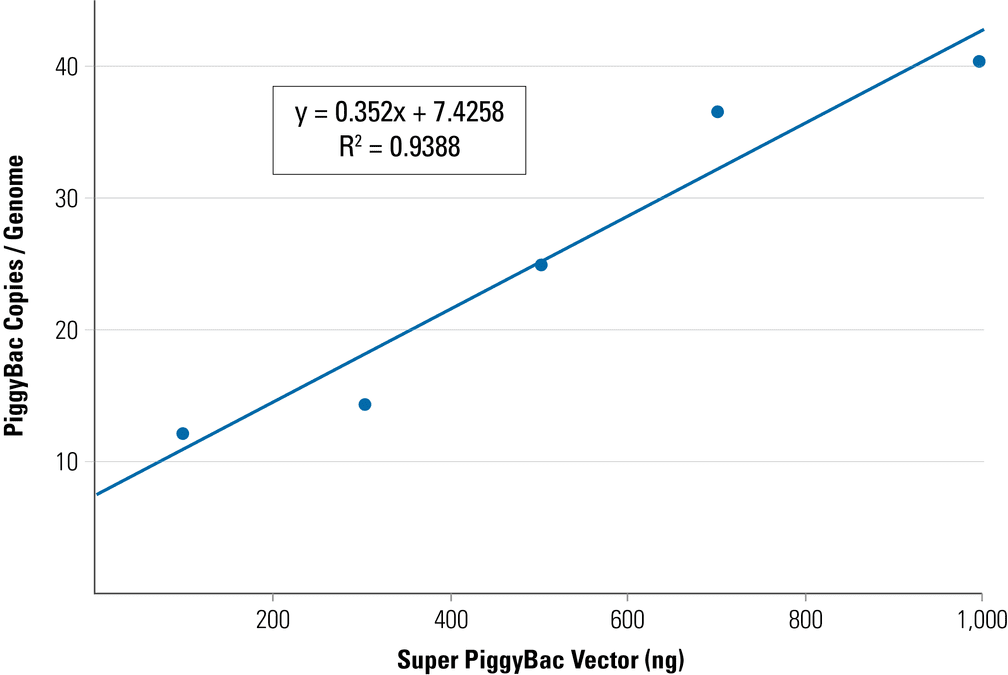
Example PiggyBac qPCR Copy Number Kit data from a PiggyBac Vector titration study—the PiggyBac insert copy number can be changed by adjusting the ratio of PiggyBac Vector to Super PiggyBac Transposase Expression Vector:
Figure 1. Adjust the PiggyBac insert copy number by changing the ratio of PiggyBac Vector to Super PiggyBac Transposase. To achieve the indicated ratio of PiggyBac Vector to Super PiggyBac Transposase Expression Vector, amount of PiggyBac Vector added was 100 ng, 300 ng, 500 ng, 700 ng, and 1,000 ng, respectively, while the amount of Super PiggyBac Transposase Expression Vector was held constant at 100 ng.
FAQs
Documentation
Citations
Related Products
Products
| Catalog Number | Description | Size | Price | Quantity | Add to Cart | |||
|---|---|---|---|---|---|---|---|---|
| PBC100A-1 | piggyBac qPCR copy number kit, 20 reactions | 20 Reactions | $460 |
|
||||
Overview
Overview
Confidently count your PiggyBac insertsWhen you need to know how many PiggyBac integration events have happened, turn to SBI’s PiggyBac qPCR Copy Number Kit. This kit uses qPCR to measure the number of PiggyBac inserts relative to a specific genomic locus—the UCR1 element—with the PiggyBac insert copy number calculated using the cycle threshold (Ct) values of the UCR1 signal relative to the PiggyBac insert signal.
The PiggyBac qPCR Copy Number Kit comes with enough reagents—UCR1 primers, PiggyBac primers, and cell lysis buffer—for twenty copy number determinations, and is compatible with all of SBI’s PiggyBac Vectors.
NOTE: Your cells must be passaged at least once before performing this copy number measurement to ensure that residual, non-integrated piggyBac transposon plasmid does not interfere with the qPCR reaction.
References
How It Works
How It Works
Calculate PiggyBac insert copy number
The PiggyBac qPCR Copy Number Kit provides robust PiggyBac insert copy number determination. To calculate the PiggyBac insert copy number from the Ct values:
- Calculate the average Cts for the PiggyBac inserts and for the UCR1 loci (there are two UCR1 elements per genome)
- The copy number is the ΔΔCt/2—the ΔΔCt value must be divided by two to account for the two UCR1 elements:
ΔΔCt = 2-((average PiggyBac insert Ct) – (average UCR1 Ct))
PiggyBac insert copy number = ΔΔCt/2
You can change the number of PiggyBac insertions by adjusting the ratio of PiggyBac Vector to Super PiggyBac Transposase Expression Vector (Cat.# PB210PA-1).
Supporting Data
Supporting Data
Robust PiggyBac insert copy number determination
Example PiggyBac qPCR Copy Number Kit data from a PiggyBac Vector titration study—the PiggyBac insert copy number can be changed by adjusting the ratio of PiggyBac Vector to Super PiggyBac Transposase Expression Vector:
Figure 1. Adjust the PiggyBac insert copy number by changing the ratio of PiggyBac Vector to Super PiggyBac Transposase. To achieve the indicated ratio of PiggyBac Vector to Super PiggyBac Transposase Expression Vector, amount of PiggyBac Vector added was 100 ng, 300 ng, 500 ng, 700 ng, and 1,000 ng, respectively, while the amount of Super PiggyBac Transposase Expression Vector was held constant at 100 ng.