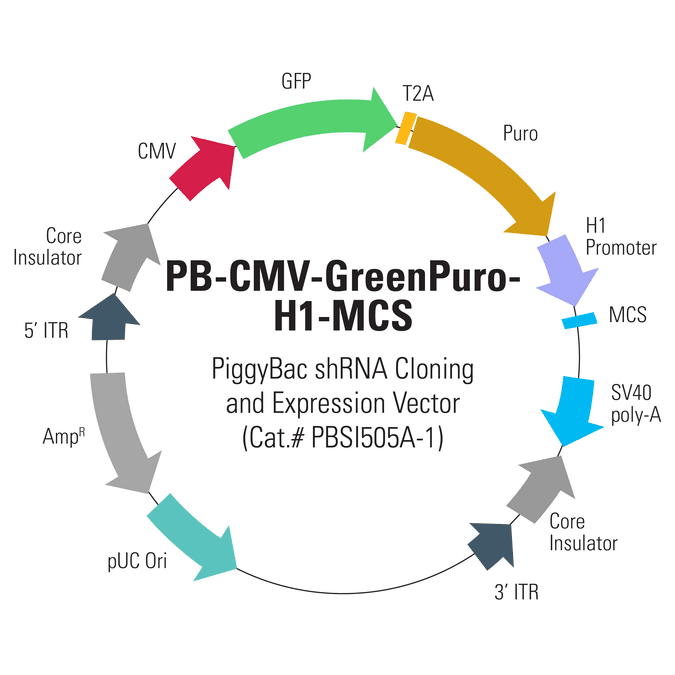
PB-CMV-GreenPuro-H1-MCS shRNA Cloning and Expression Vector
- Make transgenic cell lines with a single transfection
- Integrate multiple PiggyBac Vectors in a single transfection
- Insert an expression cassette into human, mouse, and rat cells
- Deliver virtually any-sized DNA insert, from 10 – 100 kb
- Choose from PiggyBac Vectors that express your gene-of-interest from constitutive or inducible promoters and include a variety of markers
Products
| Catalog Number | Description | Size | Price | Quantity | Add to Cart | |||
|---|---|---|---|---|---|---|---|---|
| PBSI505A-1 | PB-CMV-GreenPuro-H1-MCS shRNA cloning and expression vector | 10 µg | $1025 |
|
||||
Overview
Overview
Easy and consistent shRNA delivery and expression
Not just for genes, the PiggyBac system is also an excellent choice for reliably producing shRNA. The PB-CMV-GreenPuro-H1-MCS shRNA Cloning and Expression Vector (Cat.# PBSI505A-1) drives production of your shRNA from the strong H1 promoter. The vector also features GFP and puromycin resistance co-expressed from the strong CMV promoter, with co-expression mediated by the T2A element.
 With the PiggyBac Transposon System, you can:
With the PiggyBac Transposon System, you can:
- Make transgenic cell lines with a single transfection
- Integrate multiple PiggyBac Vectors in a single transfection
- Insert an expression cassette into human, mouse, and rat cells
- Deliver virtually any-sized DNA insert, from 10 – 100 kb
- Choose from PiggyBac Vectors that express your gene-of-interest from constitutive or inducible promoters and include a variety of markers
- Determine the number of integration events with the PiggyBac qPCR Copy Number Kit (# PBC100A-1)
Customer Agreements
Academic customers can purchase PiggyBac Transposon System components for internal research purposes for indefinite use, whereas commercial customers must sign a customer agreement for a six-month, limited-use license to evaluate the technology.
For end user license information, see the following:
* SBI is fully licensed to distribute PiggyBac vectors as a partnership with Hera BioLabs, Inc.
References
How It Works
How It Works
The PiggyBac Transposon System’s Cut-and-Paste Mechanism
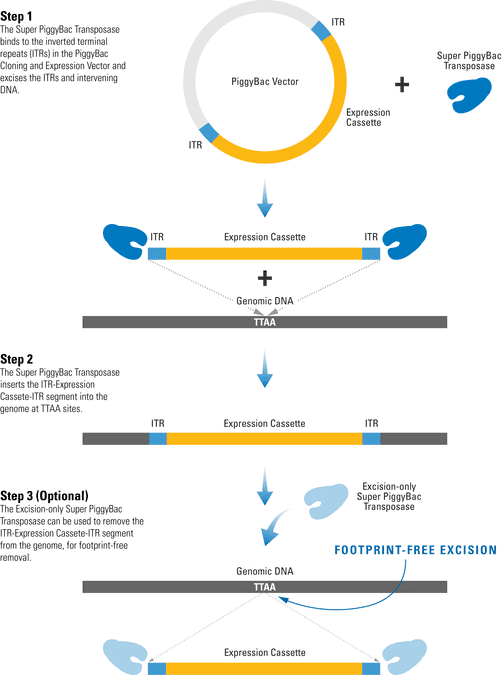
The efficient PiggyBac Transposon System uses a cut-and-paste mechanism to transfer DNA from the PiggyBac Vector into the genome. If only temporary genomic integration is desired, the Excision-only PiggyBac Transposase can be transiently expressed for footprint-free removal of the insert, resulting in reconstitution of the original genome sequence.
Figure 1. The PiggyBac Transposon System’s cut-and-paste mechanism.
- The Super PiggyBac Transposase binds to specific inverted terminal repeats (ITRs) in the PiggyBac Cloning and Expression Vector and excises the ITRs and intervening DNA.
- The Super PiggyBac Transposase inserts the ITR-Expression Cassette-ITR segment into the genome at TTAA sites.
- The Excision-only Super PiggyBac Transposase can be used to remove the ITR-Expression Cassette-ITR segment from the genome, for footprint-free removal
Supporting Data
FAQs
Documentation
Citations
Related Products
Products
| Catalog Number | Description | Size | Price | Quantity | Add to Cart | |||
|---|---|---|---|---|---|---|---|---|
| PBSI505A-1 | PB-CMV-GreenPuro-H1-MCS shRNA cloning and expression vector | 10 µg | $1025 |
|
||||
Overview
Overview
Easy and consistent shRNA delivery and expression
Not just for genes, the PiggyBac system is also an excellent choice for reliably producing shRNA. The PB-CMV-GreenPuro-H1-MCS shRNA Cloning and Expression Vector (Cat.# PBSI505A-1) drives production of your shRNA from the strong H1 promoter. The vector also features GFP and puromycin resistance co-expressed from the strong CMV promoter, with co-expression mediated by the T2A element.
 With the PiggyBac Transposon System, you can:
With the PiggyBac Transposon System, you can:
- Make transgenic cell lines with a single transfection
- Integrate multiple PiggyBac Vectors in a single transfection
- Insert an expression cassette into human, mouse, and rat cells
- Deliver virtually any-sized DNA insert, from 10 – 100 kb
- Choose from PiggyBac Vectors that express your gene-of-interest from constitutive or inducible promoters and include a variety of markers
- Determine the number of integration events with the PiggyBac qPCR Copy Number Kit (# PBC100A-1)
Customer Agreements
Academic customers can purchase PiggyBac Transposon System components for internal research purposes for indefinite use, whereas commercial customers must sign a customer agreement for a six-month, limited-use license to evaluate the technology.
For end user license information, see the following:
* SBI is fully licensed to distribute PiggyBac vectors as a partnership with Hera BioLabs, Inc.
References
How It Works
How It Works
The PiggyBac Transposon System’s Cut-and-Paste Mechanism
The efficient PiggyBac Transposon System uses a cut-and-paste mechanism to transfer DNA from the PiggyBac Vector into the genome. If only temporary genomic integration is desired, the Excision-only PiggyBac Transposase can be transiently expressed for footprint-free removal of the insert, resulting in reconstitution of the original genome sequence.
Figure 1. The PiggyBac Transposon System’s cut-and-paste mechanism.
- The Super PiggyBac Transposase binds to specific inverted terminal repeats (ITRs) in the PiggyBac Cloning and Expression Vector and excises the ITRs and intervening DNA.
- The Super PiggyBac Transposase inserts the ITR-Expression Cassette-ITR segment into the genome at TTAA sites.
- The Excision-only Super PiggyBac Transposase can be used to remove the ITR-Expression Cassette-ITR segment from the genome, for footprint-free removal