PB-EF1α-MCS-IRES-Neo PiggyBac cDNA Cloning and Expression Vector
- Make transgenic cell lines with a single transfection
- Integrate multiple PiggyBac Vectors in a single transfection
- Insert an expression cassette into human, mouse, and rat cells
- Deliver virtually any-sized DNA insert, from 10 – 100 kb
- Choose from PiggyBac Vectors that express your gene-of-interest from constitutive or inducible promoters and include a variety of markers
Products
| Catalog Number | Description | Size | Price | Quantity | Add to Cart | |||
|---|---|---|---|---|---|---|---|---|
| PB533A-2 | PB-EF1α-MCS-IRES-Neo cDNA cloning and expression vector | 10 µg | $1025 |
|
||||
Overview
Overview
Get easy, consistent transgenesis
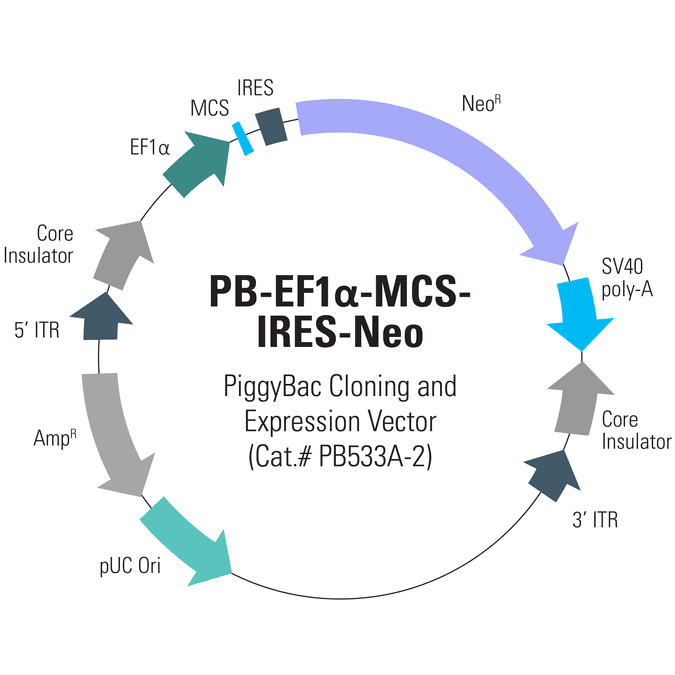
Consistent and easy-to-use, SBI’s PiggyBac Transposon System includes cloning and expression vectors that come with a range of markers as well as both constitutive and inducible promoters. The PB-EF1α-MCS-IRES-Neo PiggyBac cDNA Cloning and Expression Vector (Cat.# PB533A-2) drives co-expression of your gene-of-interest and the neomycin marker from the moderate EF1α promoter. Co-expression is mediated by an IRES element upstream of the neomycin resistance gene.
 Why use the PiggyBac Transposon System?
Why use the PiggyBac Transposon System?
Easy, consistent transgenesis with no limits on cargo size—For transgenesis that’s easy, consistent, and not limited by cargo size, SBI’s PiggyBac Transposon System is an excellent choice. The system consists of a PiggyBac Vector and the Super PiggyBac Transposase which recognizes transposon-specific inverted terminal repeats (ITRs) and efficiently integrates the ITRs and intervening DNA into the genome at TTAA sites. The Super PiggyBac Transposase is delivered to the cell via the Super PiggyBac Transposase Expression Vector, which is co-transfected with one or more PiggyBac Vectors.
Footprint-free removal that leaves no PiggyBac sequences behind—In addition to ease-of-use, consistency, and the lack of limits on DNA insert size, what sets this system apart is the ability to reverse the integration reaction in a footprint-free way—with the Excision Only PiggyBac Transposase (Cat.# PB220PA-1), the ITRs and cargo that the Super PiggyBac Transposase integrates into the genome can be removed, leaving behind the original genomic sequence and nothing else.
- Make transgenic cell lines with a single transfection
- Integrate multiple PiggyBac Vectors in a single transfection
- Insert an expression cassette into human, mouse, and rat cells
- Deliver virtually any-sized DNA insert, from 10 – 100 kb
- Choose from PiggyBac Vectors that express your gene-of-interest from constitutive or inducible promoters and include a variety of markers
- Determine the number of integration events with the PiggyBac qPCR Copy Number Kit (# PBC100A-1)
Customer Agreements
Academic customers can purchase PiggyBac Transposon System components for internal research purposes for indefinite use, whereas commercial customers must sign a customer agreement for a six-month, limited-use license to evaluate the technology.
* SBI is fully licensed to distribute PiggyBac vectors as a partnership with Hera BioLabs, Inc.
References
How It Works
How It Works
The PiggyBac Transposon System’s Cut-and-Paste Mechanism
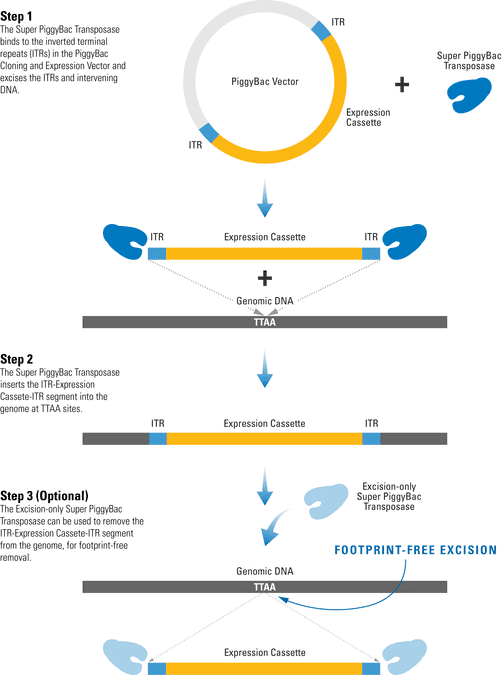
The efficient PiggyBac Transposon System uses a cut-and-paste mechanism to transfer DNA from the PiggyBac Vector into the genome. If only temporary genomic integration is desired, the Excision-only PiggyBac Transposase can be transiently expressed for footprint-free removal of the insert, resulting in reconstitution of the original genome sequence.
Figure 1. The PiggyBac Transposon System’s cut-and-paste mechanism.
- The Super PiggyBac Transposase binds to specific inverted terminal repeats (ITRs) in the PiggyBac Cloning and Expression Vector and excises the ITRs and intervening DNA.
- The Super PiggyBac Transposase inserts the ITR-Expression Cassette-ITR segment into the genome at TTAA sites.
- The Excision-only Super PiggyBac Transposase can be used to remove the ITR-Expression Cassette-ITR segment from the genome, for footprint-free removal
Supporting Data
Supporting Data
One transfection can integrate one or more genes that can be precisely removed
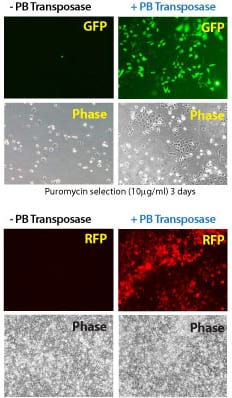
Figure 2. Efficient transgenesis with the Super PiggyBac Transposase and both single- and dual-promoter PiggyBac Vectors. (Top four panels) Co-transfection with the Super PiggyBac Transposase Expression Vector (Cat.# PB210PA-1) and a Dual Promoter PiggyBac Cloning and Expression Vector (Cat.# PB513B-1) into HeLa cells demonstrates the efficient integration delivered by SBI’s PiggyBac Transposon System. After ten days of puromycin selection, only the cells co-transfected with the Super PiggyBac Transposase (+PB, right two panels) show robust growth and GFP fluorescence. (Bottom four panels) Co-transfection with the Super PiggyBac Transposase Expression Vector (Cat.# PB210PA-1) and a Single Promoter PiggyBac Cloning and Expression Vector (Cat.# PB531A-2) into HEK293 cells further demonstrates the efficient integration delivered by SBI’s PiggyBac Transposon System. After seven days of growth, the majority of cells that received the Super PiggyBac Transposase Expression Vector (+PB, right two panels) were RFP positive.
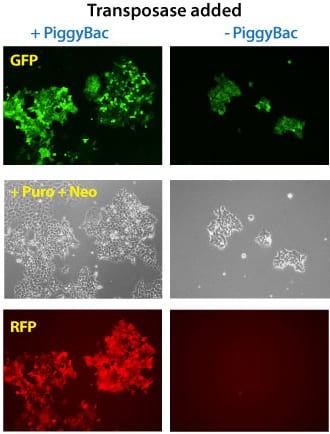
Figure 3. Simultaneous integration of multiple PiggyBac Vectors is also highly efficient. METHODS: Three different PiggyBac transposon vectors (Cat.# PB513B-1, Cat.# PB533A-2, and Cat.# PB531A-2) were co-transfected with (left panels) or without (right panels) the Super PiggyBac Transposase Expression Vector (Cat.# PB210PA-1) into Human HT1080 cells. Puromycin and neomycin selection was applied for seven days. The cells that were co-transfected with the Super PiggyBac Transposase Expression Vector were puro and neo resistant, GFP-positive, and RFP-positive. Background GFP-positive cells that are puro resistant stem from random PB513B-1 integrations during the puromycin selection. The non-PiggyBac-mediated integration rate in those cells was extremely low and no RFP-positive cells were identified.
FAQs
Documentation
Citations
Related Products
Products
| Catalog Number | Description | Size | Price | Quantity | Add to Cart | |||
|---|---|---|---|---|---|---|---|---|
| PB533A-2 | PB-EF1α-MCS-IRES-Neo cDNA cloning and expression vector | 10 µg | $1025 |
|
||||
Overview
Overview
Get easy, consistent transgenesis
Consistent and easy-to-use, SBI’s PiggyBac Transposon System includes cloning and expression vectors that come with a range of markers as well as both constitutive and inducible promoters. The PB-EF1α-MCS-IRES-Neo PiggyBac cDNA Cloning and Expression Vector (Cat.# PB533A-2) drives co-expression of your gene-of-interest and the neomycin marker from the moderate EF1α promoter. Co-expression is mediated by an IRES element upstream of the neomycin resistance gene.
 Why use the PiggyBac Transposon System?
Why use the PiggyBac Transposon System?
Easy, consistent transgenesis with no limits on cargo size—For transgenesis that’s easy, consistent, and not limited by cargo size, SBI’s PiggyBac Transposon System is an excellent choice. The system consists of a PiggyBac Vector and the Super PiggyBac Transposase which recognizes transposon-specific inverted terminal repeats (ITRs) and efficiently integrates the ITRs and intervening DNA into the genome at TTAA sites. The Super PiggyBac Transposase is delivered to the cell via the Super PiggyBac Transposase Expression Vector, which is co-transfected with one or more PiggyBac Vectors.
Footprint-free removal that leaves no PiggyBac sequences behind—In addition to ease-of-use, consistency, and the lack of limits on DNA insert size, what sets this system apart is the ability to reverse the integration reaction in a footprint-free way—with the Excision Only PiggyBac Transposase (Cat.# PB220PA-1), the ITRs and cargo that the Super PiggyBac Transposase integrates into the genome can be removed, leaving behind the original genomic sequence and nothing else.
- Make transgenic cell lines with a single transfection
- Integrate multiple PiggyBac Vectors in a single transfection
- Insert an expression cassette into human, mouse, and rat cells
- Deliver virtually any-sized DNA insert, from 10 – 100 kb
- Choose from PiggyBac Vectors that express your gene-of-interest from constitutive or inducible promoters and include a variety of markers
- Determine the number of integration events with the PiggyBac qPCR Copy Number Kit (# PBC100A-1)
Customer Agreements
Academic customers can purchase PiggyBac Transposon System components for internal research purposes for indefinite use, whereas commercial customers must sign a customer agreement for a six-month, limited-use license to evaluate the technology.
* SBI is fully licensed to distribute PiggyBac vectors as a partnership with Hera BioLabs, Inc.
References
How It Works
How It Works
The PiggyBac Transposon System’s Cut-and-Paste Mechanism
The efficient PiggyBac Transposon System uses a cut-and-paste mechanism to transfer DNA from the PiggyBac Vector into the genome. If only temporary genomic integration is desired, the Excision-only PiggyBac Transposase can be transiently expressed for footprint-free removal of the insert, resulting in reconstitution of the original genome sequence.
Figure 1. The PiggyBac Transposon System’s cut-and-paste mechanism.
- The Super PiggyBac Transposase binds to specific inverted terminal repeats (ITRs) in the PiggyBac Cloning and Expression Vector and excises the ITRs and intervening DNA.
- The Super PiggyBac Transposase inserts the ITR-Expression Cassette-ITR segment into the genome at TTAA sites.
- The Excision-only Super PiggyBac Transposase can be used to remove the ITR-Expression Cassette-ITR segment from the genome, for footprint-free removal
Supporting Data
Supporting Data
One transfection can integrate one or more genes that can be precisely removed
Figure 2. Efficient transgenesis with the Super PiggyBac Transposase and both single- and dual-promoter PiggyBac Vectors. (Top four panels) Co-transfection with the Super PiggyBac Transposase Expression Vector (Cat.# PB210PA-1) and a Dual Promoter PiggyBac Cloning and Expression Vector (Cat.# PB513B-1) into HeLa cells demonstrates the efficient integration delivered by SBI’s PiggyBac Transposon System. After ten days of puromycin selection, only the cells co-transfected with the Super PiggyBac Transposase (+PB, right two panels) show robust growth and GFP fluorescence. (Bottom four panels) Co-transfection with the Super PiggyBac Transposase Expression Vector (Cat.# PB210PA-1) and a Single Promoter PiggyBac Cloning and Expression Vector (Cat.# PB531A-2) into HEK293 cells further demonstrates the efficient integration delivered by SBI’s PiggyBac Transposon System. After seven days of growth, the majority of cells that received the Super PiggyBac Transposase Expression Vector (+PB, right two panels) were RFP positive.
Figure 3. Simultaneous integration of multiple PiggyBac Vectors is also highly efficient. METHODS: Three different PiggyBac transposon vectors (Cat.# PB513B-1, Cat.# PB533A-2, and Cat.# PB531A-2) were co-transfected with (left panels) or without (right panels) the Super PiggyBac Transposase Expression Vector (Cat.# PB210PA-1) into Human HT1080 cells. Puromycin and neomycin selection was applied for seven days. The cells that were co-transfected with the Super PiggyBac Transposase Expression Vector were puro and neo resistant, GFP-positive, and RFP-positive. Background GFP-positive cells that are puro resistant stem from random PB513B-1 integrations during the puromycin selection. The non-PiggyBac-mediated integration rate in those cells was extremely low and no RFP-positive cells were identified.