Have you seen the new CLOuD9 system for site-specific chromatin loop formation?
We hope you’ve seen our latest CRISPR/Cas9 product, which demonstrates one of the innovative ways scientists are starting to harness the power of CRISPR/Cas9 for easy site-specific DNA targeting. We’re especially proud of this product as it’s the result of an international, multi-site effort that included SBI scientists Fangting Wu and Chiao-Chain Huang1.
With the CLOuD9 System, you can direct chromatin loop formation at the sites you determine, enabling studies of the dynamic effects of chromatin looping on gene expression regulation, as well as other processes affected by chromatin structure.
How it works
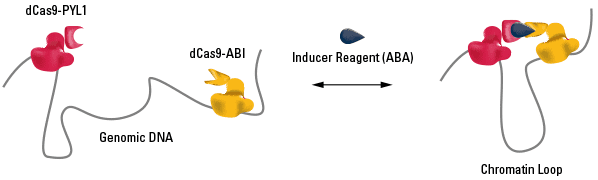
Figure 1. The CLOuD9 System enables formation of inducible and reversible chromatin loops anywhere in the genome.
The CLOuD9 System, short for Chromatin Loop Reorganization using CRISPR-dCas9, lets researchers form chromatin loops at specific locations, thanks to CRISPR/Cas9-based targeting. The CRISPR/Cas9 part of the system takes advantage of gRNA-targeted DNA binding by two different Cas9 null mutants. The null mutants have no DNA scission activity, turning Cas9 into a site-specific DNA binding protein, where the binding sites are specified by the gRNA. The CLOuD9 System uses two different Cas9 null mutants, one from Staphylococcus aureus and the other from S. pyogenes, so that two different gRNAs can be used.
The other part of the CLOuD9 System leverages the inducible protein-protein binding of the dimerization domains from ABI and PYL1, two proteins from the plant abscisic acid (ABA) signaling pathway. The dimerization domains from these proteins reversibly bind in the presence of abscisic acid—increasing abscisic acid leads to more ABI-PYL1 dimers, and washout of the abscisic acid reduces the number of ABI-PYL1 dimers.
The CLOuD9 System fuses the ABI dimerization domain to the S. aureus Cas9 null mutant and the PYL1 dimerization domain to the S. pyogenes Cas9 null mutant. The result is gRNA-directed binding of each fusion protein to a specific site in the genome, dimerization of ABI-PYL1 in the presence of abscisic acids and the ABA proteins, and, thus, formation of a chromatin loop between the S. aureus and S. pyogenes Cas9 null mutant proteins.
In some systems, the loop formation can facilitate gene expression.
See the data and learn more about the system at the CLOuD9 Gene Expression Regulation Kit product page.
References
- Morgan, SL, et al. Manipulation of nuclear architecture through CRISPR-mediated chromosomal looping. Nat Commun. 2017. Jul 13; 8:15993. PMCID: PMC5511349.