Excision-only PiggyBac Transposase Expression Vector
- Make transgenic cell lines with a single transfection
- Integrate multiple PiggyBac Vectors in a single transfection
- Insert an expression cassette into human, mouse, and rat cells
- Deliver virtually any-sized DNA insert, from 10 – 100 kb
- Choose from PiggyBac Vectors that express your gene-of-interest from constitutive or inducible promoters and include a variety of markers
Products
| Catalog Number | Description | Size | Price | Quantity | Add to Cart | |||
|---|---|---|---|---|---|---|---|---|
| PB220PA-1 | Excision only piggyBac Transposase expression vector | 10 µg | $488 |
|
||||
Overview
Overview
Reversible transgenesis that leaves no trace behind
One of the advantages of the PiggyBac Transposon System* is the reversibility of the integration event. With the Excision-only PiggyBac Transposase, delivered by transient transfection of the Exicison-only PiggyBac Transposase Expression Vector, you can remove any DNA integrated into the genome using the PiggyBac Transposon System. After removal of the PiggyBac insert, the genomic DNA is restored to the original sequence with no residual PiggyBac sequences left behind, resulting in a truly footprint-free removal.
Both the Excision-only PiggyBac Transposase and the Super PiggyBac Transposase (Cat.# PB210PA-1) recognize transposon-specific inverted terminal repeats (ITRs). While the Super PiggyBac Transposase has both excision and integration activities that enable the enzyme to mediate removal of the ITRs and intervening DNA from one location (the vector) and insert that DNA fragment into another location (the genome), the Excision-only PiggyBac Transposase only has the excision activity. Thus, the Excision-only PiggyBac Transposase can be used to remove DNA segments from the genome that are bordered by the PiggyBac transposon ITRs.
With the PiggyBac Transposon System, you can:
- Make transgenic cell lines with a single transfection
- Integrate multiple PiggyBac Vectors in a single transfection
- Insert an expression cassette into human, mouse, and rat cells
- Deliver virtually any-sized DNA insert, from 10 – 100 kb
- Choose from PiggyBac Vectors that express your gene-of-interest from constitutive or inducible promoters and include a variety of markers
- Determine the number of integration events with the PiggyBac qPCR Copy Number Kit (# PBC100A-1)
Customer Agreements
Academic customers can purchase PiggyBac Transposon System components for internal research purposes for indefinite use, whereas commercial customers must sign a customer agreement for a six-month, limited-use license to evaluate the technology.
* SBI is fully licensed to distribute PiggyBac vectors as a partnership with Hera BioLabs, Inc.
How It Works
How It Works
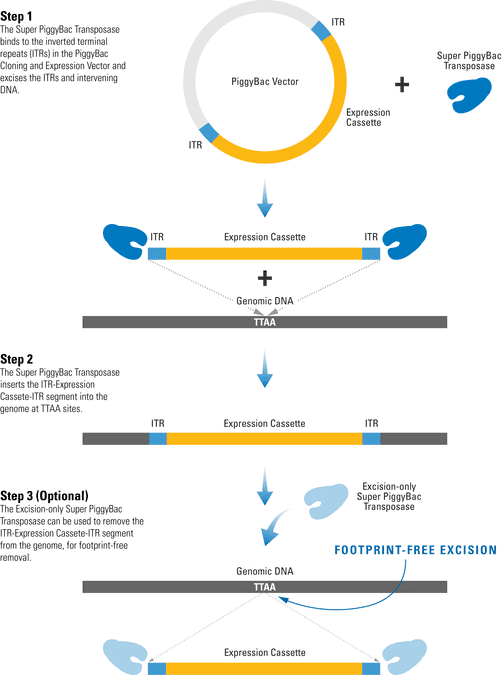
The PiggyBac Transposon System’s Cut-and-Paste Mechanism
The efficient PiggyBac Transposon System uses a cut-and-paste mechanism to transfer DNA from the PiggyBac Vector into the genome. If only temporary genomic integration is desired, the Excision-only PiggyBac Transposase can be transiently expressed for footprint-free removal of the insert, resulting in reconstitution of the original genome sequence.
Figure 1. The PiggyBac Transposon System’s cut-and-paste mechanism.
- The Super PiggyBac Transposase binds to specific inverted terminal repeats (ITRs) in the PiggyBac Cloning and Expression Vector and excises the ITRs and intervening DNA.
- The Super PiggyBac Transposase inserts the ITR-Expression Cassette-ITR segment into the genome at TTAA sites.
- The Excision-only Super PiggyBac Transposase can be used to remove the ITR-Expression Cassette-ITR segment from the genome, for footprint-free removal
Supporting Data
Supporting Data
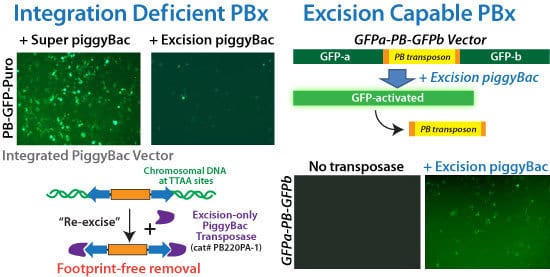
Reverse transgenesis with the PiggyBac Transposon System’s footprint-free excision process
Figure 2. SBI’s Excision-only PiggyBac Transposase is integrase-deficient but excision capable for reversible transgenesis. (Left panels) Comparison of the integration abilities of the Super PiggyBac Transposase (Cat.# PB210PA-1) to the Excision-only PiggyBac Transposase upon co-transfection with a GFP-expressing PiggyBac Vector. The very low number of GFP-positive cells in the sample co-transfected with Excision-only Transposase versus the numerous GFP-positive cells in the sample co-transfected with the Super PiggyBac Transposase demonstrate the poor integration abilities of the Excision-only enzyme. (Right panels) We inserted a GFP expression cassette into the genome, and then inactivated GFP expression by disrupting the open reading frame with a PiggyBac transposon. Successful use of the Excision-only PiggyBac Transposase restores GFP function, leading to GFP-positive cells. Restoration of GFP fluorescence also demonstrates the seamlessness of excision—the GFP coding region would remain disrupted if the transposon sequences did not get completely removed.
FAQs
Resources
Related Products
Citations
-
Brouwer, I, de Kort, MAC & Lenstra, TL. (2024) Measuring Transcription Dynamics of Individual Genes Inside Living Cells. Methods in molecular biology (Clifton, N.J.). 2024; 2694:235-265. PM ID: 37824008
-
Matta, SK, et al. (2024) Genome-wide and targeted CRISPR screens identify RNF213 as a mediator of interferon gamma-dependent pathogen restriction in human cells. Proceedings of the National Academy of Sciences of the United States of America. 2024; 121(1):e2315865120. PM ID: 38147552
-
Cho, MG, et al. (2024) MRE11 liberates cGAS from nucleosome sequestration during tumorigenesis. Nature. 2024; 625(7995):585-592. PM ID: 38200309
-
Du, M, et al. (2024) Direct observation of a condensate effect on super-enhancer controlled gene bursting. Cell. 2024; 187(2):331-344.e17. PM ID: 38194964
-
Schmitt, J, et al. (2024) Repurposing an endogenous degradation domain for antibody-mediated disposal of cell-surface proteins. EMBO reports. 2024;. PM ID: 38287192
-
Byrnes, AE, et al. (2024) A fluorescent splice-switching mouse model enables high-throughput, sensitive quantification of antisense oligonucleotide delivery and activity. Cell reports methods. 2024; 4(1):100673. PM ID: 38171361
-
Daiki, K, et al. (2024) Blood Endocan as a Biomarker for Breast Cancer Recurrence. Preprint. 2024;. Link: Preprint
-
Koeppel, J, et al. (2024) Randomizing the human genome by engineering recombination between repeat elements. bioRxiv. 2024;. Link: bioRxiv
-
Kortleve, D, et al. (2024) TCR-engineered T-cells directed against Ropporin-1 constitute a safe and effective treatment for triple-negative breast cancer in near-clinical models. bioRxiv. 2024;. Link: bioRxiv
-
Haakonsen, DL, et al. (2024) Stress response silencing by an E3 ligase mutated in neurodegeneration. Nature. 2024; 626(8000):874-880. PM ID: 38297121
-
Gupta, P, et al. (2024) Development of pathophysiologically relevant models of sickle cell disease and β-thalassemia for therapeutic studies. Nature communications. 2024; 15(1):1794. PM ID: 38413594
-
Company, C, et al. (2024) Logical design of synthetic cis-regulatory DNA for genetic tracing of cell identities and state changes. Nature communications. 2024; 15(1):897. PM ID: 38316783
-
Yang, L, et al. (2024) Uncovering receptor-ligand interactions using a high-avidity CRISPR activation screening platform. Science advances. 2024; 10(7):eadj2445. PM ID: 38354234
-
Kubara, K, et al. (2024) Lymph node macrophages drive innate immune responses to enhance the anti-tumor efficacy of mRNA vaccines. Molecular therapy : the journal of the American Society of Gene Therapy. 2024;. PM ID: 38243602
-
Ng-Blichfeldt, J, et al. (2024) Identification of a core transcriptional program driving the human renal mesenchymal-to-epithelial transition. Developmental Cell. 2024;. Link: Developmental Cell
-
Yang, J, Cook, L & Chen, Z. (2024) Systematic evaluation of retroviral LTRs as cis-regulatory elements in mouse embryos. Cell reports. 2024; 43(3):113775. PM ID: 38381606
-
Taglini, F, et al. (2024) DNMT3B PWWP mutations cause hypermethylation of heterochromatin. EMBO reports. 2024;. PM ID: 38291337
-
Tanase-Nakao, K, et al. (2024) Genotype-Phenotype Correlations in Thirty Japanese Patients with Congenital Hypothyroidism Attributable to TG Defects. The Journal of clinical endocrinology and metabolism. 2024;. PM ID: 38373250
-
Alsouri, S, et al. (2024) Actinin-4 controls survival signaling in B cells by limiting the lateral mobility of B-cell antigen receptors. European journal of immunology. 2024;:e2350774. PM ID: 38299456
-
Ke, X, et al. (2024) Establishment of a novel minigenome system for the identification of drugs targeting Nipah virus replication. The Journal of general virology. 2024; 105(1). PM ID: 38180473
- See More
Products
| Catalog Number | Description | Size | Price | Quantity | Add to Cart | |||
|---|---|---|---|---|---|---|---|---|
| PB220PA-1 | Excision only piggyBac Transposase expression vector | 10 µg | $488 |
|
||||
Overview
Overview
Reversible transgenesis that leaves no trace behind
One of the advantages of the PiggyBac Transposon System* is the reversibility of the integration event. With the Excision-only PiggyBac Transposase, delivered by transient transfection of the Exicison-only PiggyBac Transposase Expression Vector, you can remove any DNA integrated into the genome using the PiggyBac Transposon System. After removal of the PiggyBac insert, the genomic DNA is restored to the original sequence with no residual PiggyBac sequences left behind, resulting in a truly footprint-free removal.
Both the Excision-only PiggyBac Transposase and the Super PiggyBac Transposase (Cat.# PB210PA-1) recognize transposon-specific inverted terminal repeats (ITRs). While the Super PiggyBac Transposase has both excision and integration activities that enable the enzyme to mediate removal of the ITRs and intervening DNA from one location (the vector) and insert that DNA fragment into another location (the genome), the Excision-only PiggyBac Transposase only has the excision activity. Thus, the Excision-only PiggyBac Transposase can be used to remove DNA segments from the genome that are bordered by the PiggyBac transposon ITRs.
With the PiggyBac Transposon System, you can:
- Make transgenic cell lines with a single transfection
- Integrate multiple PiggyBac Vectors in a single transfection
- Insert an expression cassette into human, mouse, and rat cells
- Deliver virtually any-sized DNA insert, from 10 – 100 kb
- Choose from PiggyBac Vectors that express your gene-of-interest from constitutive or inducible promoters and include a variety of markers
- Determine the number of integration events with the PiggyBac qPCR Copy Number Kit (# PBC100A-1)
Customer Agreements
Academic customers can purchase PiggyBac Transposon System components for internal research purposes for indefinite use, whereas commercial customers must sign a customer agreement for a six-month, limited-use license to evaluate the technology.
* SBI is fully licensed to distribute PiggyBac vectors as a partnership with Hera BioLabs, Inc.
How It Works
How It Works
The PiggyBac Transposon System’s Cut-and-Paste Mechanism
The efficient PiggyBac Transposon System uses a cut-and-paste mechanism to transfer DNA from the PiggyBac Vector into the genome. If only temporary genomic integration is desired, the Excision-only PiggyBac Transposase can be transiently expressed for footprint-free removal of the insert, resulting in reconstitution of the original genome sequence.
Figure 1. The PiggyBac Transposon System’s cut-and-paste mechanism.
- The Super PiggyBac Transposase binds to specific inverted terminal repeats (ITRs) in the PiggyBac Cloning and Expression Vector and excises the ITRs and intervening DNA.
- The Super PiggyBac Transposase inserts the ITR-Expression Cassette-ITR segment into the genome at TTAA sites.
- The Excision-only Super PiggyBac Transposase can be used to remove the ITR-Expression Cassette-ITR segment from the genome, for footprint-free removal
Supporting Data
Supporting Data
Reverse transgenesis with the PiggyBac Transposon System’s footprint-free excision process
Figure 2. SBI’s Excision-only PiggyBac Transposase is integrase-deficient but excision capable for reversible transgenesis. (Left panels) Comparison of the integration abilities of the Super PiggyBac Transposase (Cat.# PB210PA-1) to the Excision-only PiggyBac Transposase upon co-transfection with a GFP-expressing PiggyBac Vector. The very low number of GFP-positive cells in the sample co-transfected with Excision-only Transposase versus the numerous GFP-positive cells in the sample co-transfected with the Super PiggyBac Transposase demonstrate the poor integration abilities of the Excision-only enzyme. (Right panels) We inserted a GFP expression cassette into the genome, and then inactivated GFP expression by disrupting the open reading frame with a PiggyBac transposon. Successful use of the Excision-only PiggyBac Transposase restores GFP function, leading to GFP-positive cells. Restoration of GFP fluorescence also demonstrates the seamlessness of excision—the GFP coding region would remain disrupted if the transposon sequences did not get completely removed.
FAQs
Citations
-
Brouwer, I, de Kort, MAC & Lenstra, TL. (2024) Measuring Transcription Dynamics of Individual Genes Inside Living Cells. Methods in molecular biology (Clifton, N.J.). 2024; 2694:235-265. PM ID: 37824008
-
Matta, SK, et al. (2024) Genome-wide and targeted CRISPR screens identify RNF213 as a mediator of interferon gamma-dependent pathogen restriction in human cells. Proceedings of the National Academy of Sciences of the United States of America. 2024; 121(1):e2315865120. PM ID: 38147552
-
Cho, MG, et al. (2024) MRE11 liberates cGAS from nucleosome sequestration during tumorigenesis. Nature. 2024; 625(7995):585-592. PM ID: 38200309
-
Du, M, et al. (2024) Direct observation of a condensate effect on super-enhancer controlled gene bursting. Cell. 2024; 187(2):331-344.e17. PM ID: 38194964
-
Schmitt, J, et al. (2024) Repurposing an endogenous degradation domain for antibody-mediated disposal of cell-surface proteins. EMBO reports. 2024;. PM ID: 38287192
-
Byrnes, AE, et al. (2024) A fluorescent splice-switching mouse model enables high-throughput, sensitive quantification of antisense oligonucleotide delivery and activity. Cell reports methods. 2024; 4(1):100673. PM ID: 38171361
-
Daiki, K, et al. (2024) Blood Endocan as a Biomarker for Breast Cancer Recurrence. Preprint. 2024;. Link: Preprint
-
Koeppel, J, et al. (2024) Randomizing the human genome by engineering recombination between repeat elements. bioRxiv. 2024;. Link: bioRxiv
-
Kortleve, D, et al. (2024) TCR-engineered T-cells directed against Ropporin-1 constitute a safe and effective treatment for triple-negative breast cancer in near-clinical models. bioRxiv. 2024;. Link: bioRxiv
-
Haakonsen, DL, et al. (2024) Stress response silencing by an E3 ligase mutated in neurodegeneration. Nature. 2024; 626(8000):874-880. PM ID: 38297121
-
Gupta, P, et al. (2024) Development of pathophysiologically relevant models of sickle cell disease and β-thalassemia for therapeutic studies. Nature communications. 2024; 15(1):1794. PM ID: 38413594
-
Company, C, et al. (2024) Logical design of synthetic cis-regulatory DNA for genetic tracing of cell identities and state changes. Nature communications. 2024; 15(1):897. PM ID: 38316783
-
Yang, L, et al. (2024) Uncovering receptor-ligand interactions using a high-avidity CRISPR activation screening platform. Science advances. 2024; 10(7):eadj2445. PM ID: 38354234
-
Kubara, K, et al. (2024) Lymph node macrophages drive innate immune responses to enhance the anti-tumor efficacy of mRNA vaccines. Molecular therapy : the journal of the American Society of Gene Therapy. 2024;. PM ID: 38243602
-
Ng-Blichfeldt, J, et al. (2024) Identification of a core transcriptional program driving the human renal mesenchymal-to-epithelial transition. Developmental Cell. 2024;. Link: Developmental Cell
-
Yang, J, Cook, L & Chen, Z. (2024) Systematic evaluation of retroviral LTRs as cis-regulatory elements in mouse embryos. Cell reports. 2024; 43(3):113775. PM ID: 38381606
-
Taglini, F, et al. (2024) DNMT3B PWWP mutations cause hypermethylation of heterochromatin. EMBO reports. 2024;. PM ID: 38291337
-
Tanase-Nakao, K, et al. (2024) Genotype-Phenotype Correlations in Thirty Japanese Patients with Congenital Hypothyroidism Attributable to TG Defects. The Journal of clinical endocrinology and metabolism. 2024;. PM ID: 38373250
-
Alsouri, S, et al. (2024) Actinin-4 controls survival signaling in B cells by limiting the lateral mobility of B-cell antigen receptors. European journal of immunology. 2024;:e2350774. PM ID: 38299456
-
Ke, X, et al. (2024) Establishment of a novel minigenome system for the identification of drugs targeting Nipah virus replication. The Journal of general virology. 2024; 105(1). PM ID: 38180473
- See More