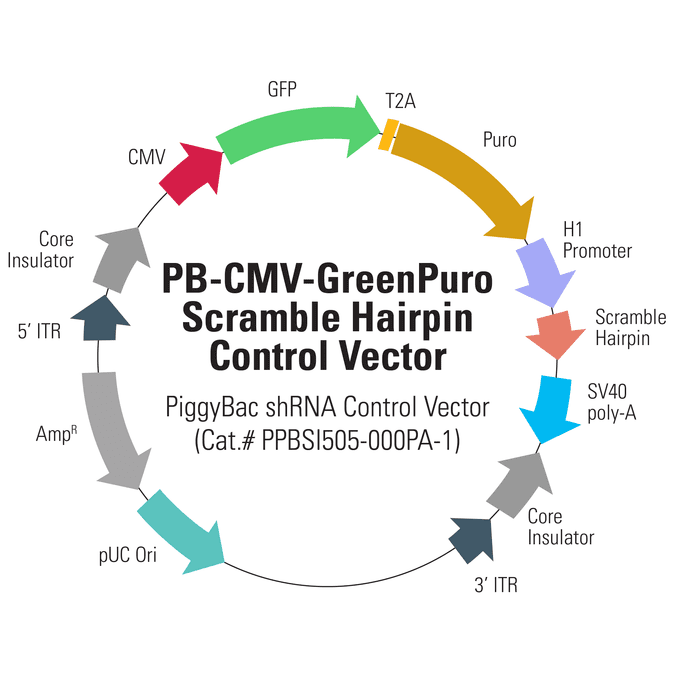
PB-CMV-GreenPuro Scramble Hairpin Control Vector
- Make transgenic cell lines with a single transfection
- Integrate multiple PiggyBac Vectors in a single transfection
- Insert an expression cassette into human, mouse, and rat cells
- Deliver virtually any-sized DNA insert, from 10 – 100 kb
- Choose from PiggyBac Vectors that express your gene-of-interest from constitutive or inducible promoters and include a variety of markers
Products
| Catalog Number | Description | Size | Price | Quantity | Add to Cart | |||
|---|---|---|---|---|---|---|---|---|
| PBSI505-000PA-1 | PB-CMV-GreenPuro Scramble Hairpin Control Vector | 10 µg | $648 |
|
||||
Overview
Overview
Easy and consistent shRNA delivery and expression
Not just for genes, the PiggyBac system is also an excellent choice for reliably producing shRNA . The PB-CMV-GreenPuro Scramble Hairpin Control Vector (Cat.# PBSI505-000PA-1) is a ready-to-transfect control for your PiggyBac shRNA projects. The vector features GFP and puromycin resistance co-expressed from the strong CMV promoter, with co-expression mediated by the T2A element. An H1 promoter drives expression of the scramble hairpin.
 With the PiggyBac Transposon System, you can:
With the PiggyBac Transposon System, you can:
- Make transgenic cell lines with a single transfection
- Integrate multiple PiggyBac Vectors in a single transfection
- Insert an expression cassette into human, mouse, and rat cells
- Deliver virtually any-sized DNA insert, from 10 – 100 kb
- Choose from PiggyBac Vectors that express your gene-of-interest from constitutive or inducible promoters and include a variety of markers
- Determine the number of integration events with the PiggyBac qPCR Copy Number Kit (# PBC100A-1)
Customer Agreements
Academic customers can purchase PiggyBac Transposon System components for internal research purposes for indefinite use, whereas commercial customers must sign a customer agreement for a six-month, limited-use license to evaluate the technology.
For end user license information, see the following:
* SBI is fully licensed to distribute PiggyBac vectors as a partnership with Hera BioLabs, Inc.
How It Works
How It Works
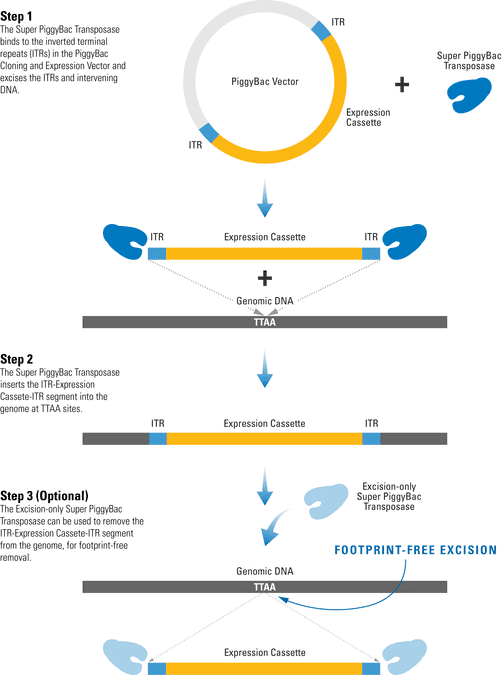
The PiggyBac Transposon System’s Cut-and-Paste Mechanism
The efficient PiggyBac Transposon System uses a cut-and-paste mechanism to transfer DNA from the PiggyBac Vector into the genome. If only temporary genomic integration is desired, the Excision-only PiggyBac Transposase can be transiently expressed for footprint-free removal of the insert, resulting in reconstitution of the original genome sequence.
Figure 1. The PiggyBac Transposon System’s cut-and-paste mechanism.
- The Super PiggyBac Transposase binds to specific inverted terminal repeats (ITRs) in the PiggyBac Cloning and Expression Vector and excises the ITRs and intervening DNA.
- The Super PiggyBac Transposase inserts the ITR-Expression Cassette-ITR segment into the genome at TTAA sites.
- The Excision-only Super PiggyBac Transposase can be used to remove the ITR-Expression Cassette-ITR segment from the genome, for footprint-free removal
Supporting Data
FAQs
Resources
Related Products
Citations
-
Brouwer, I, de Kort, MAC & Lenstra, TL. (2024) Measuring Transcription Dynamics of Individual Genes Inside Living Cells. Methods in molecular biology (Clifton, N.J.). 2024; 2694:235-265. PM ID: 37824008
-
Matta, SK, et al. (2024) Genome-wide and targeted CRISPR screens identify RNF213 as a mediator of interferon gamma-dependent pathogen restriction in human cells. Proceedings of the National Academy of Sciences of the United States of America. 2024; 121(1):e2315865120. PM ID: 38147552
-
Cho, MG, et al. (2024) MRE11 liberates cGAS from nucleosome sequestration during tumorigenesis. Nature. 2024; 625(7995):585-592. PM ID: 38200309
-
Du, M, et al. (2024) Direct observation of a condensate effect on super-enhancer controlled gene bursting. Cell. 2024; 187(2):331-344.e17. PM ID: 38194964
-
Schmitt, J, et al. (2024) Repurposing an endogenous degradation domain for antibody-mediated disposal of cell-surface proteins. EMBO reports. 2024;. PM ID: 38287192
-
Byrnes, AE, et al. (2024) A fluorescent splice-switching mouse model enables high-throughput, sensitive quantification of antisense oligonucleotide delivery and activity. Cell reports methods. 2024; 4(1):100673. PM ID: 38171361
-
Daiki, K, et al. (2024) Blood Endocan as a Biomarker for Breast Cancer Recurrence. Preprint. 2024;. Link: Preprint
-
Koeppel, J, et al. (2024) Randomizing the human genome by engineering recombination between repeat elements. bioRxiv. 2024;. Link: bioRxiv
-
Kortleve, D, et al. (2024) TCR-engineered T-cells directed against Ropporin-1 constitute a safe and effective treatment for triple-negative breast cancer in near-clinical models. bioRxiv. 2024;. Link: bioRxiv
-
Haakonsen, DL, et al. (2024) Stress response silencing by an E3 ligase mutated in neurodegeneration. Nature. 2024; 626(8000):874-880. PM ID: 38297121
-
Gupta, P, et al. (2024) Development of pathophysiologically relevant models of sickle cell disease and β-thalassemia for therapeutic studies. Nature communications. 2024; 15(1):1794. PM ID: 38413594
-
Company, C, et al. (2024) Logical design of synthetic cis-regulatory DNA for genetic tracing of cell identities and state changes. Nature communications. 2024; 15(1):897. PM ID: 38316783
-
Yang, L, et al. (2024) Uncovering receptor-ligand interactions using a high-avidity CRISPR activation screening platform. Science advances. 2024; 10(7):eadj2445. PM ID: 38354234
-
Kubara, K, et al. (2024) Lymph node macrophages drive innate immune responses to enhance the anti-tumor efficacy of mRNA vaccines. Molecular therapy : the journal of the American Society of Gene Therapy. 2024;. PM ID: 38243602
-
Ng-Blichfeldt, J, et al. (2024) Identification of a core transcriptional program driving the human renal mesenchymal-to-epithelial transition. Developmental Cell. 2024;. Link: Developmental Cell
-
Yang, J, Cook, L & Chen, Z. (2024) Systematic evaluation of retroviral LTRs as cis-regulatory elements in mouse embryos. Cell reports. 2024; 43(3):113775. PM ID: 38381606
-
Taglini, F, et al. (2024) DNMT3B PWWP mutations cause hypermethylation of heterochromatin. EMBO reports. 2024;. PM ID: 38291337
-
Tanase-Nakao, K, et al. (2024) Genotype-Phenotype Correlations in Thirty Japanese Patients with Congenital Hypothyroidism Attributable to TG Defects. The Journal of clinical endocrinology and metabolism. 2024;. PM ID: 38373250
-
Alsouri, S, et al. (2024) Actinin-4 controls survival signaling in B cells by limiting the lateral mobility of B-cell antigen receptors. European journal of immunology. 2024;:e2350774. PM ID: 38299456
-
Ke, X, et al. (2024) Establishment of a novel minigenome system for the identification of drugs targeting Nipah virus replication. The Journal of general virology. 2024; 105(1). PM ID: 38180473
- See More
Products
| Catalog Number | Description | Size | Price | Quantity | Add to Cart | |||
|---|---|---|---|---|---|---|---|---|
| PBSI505-000PA-1 | PB-CMV-GreenPuro Scramble Hairpin Control Vector | 10 µg | $648 |
|
||||
Overview
Overview
Easy and consistent shRNA delivery and expression
Not just for genes, the PiggyBac system is also an excellent choice for reliably producing shRNA . The PB-CMV-GreenPuro Scramble Hairpin Control Vector (Cat.# PBSI505-000PA-1) is a ready-to-transfect control for your PiggyBac shRNA projects. The vector features GFP and puromycin resistance co-expressed from the strong CMV promoter, with co-expression mediated by the T2A element. An H1 promoter drives expression of the scramble hairpin.
 With the PiggyBac Transposon System, you can:
With the PiggyBac Transposon System, you can:
- Make transgenic cell lines with a single transfection
- Integrate multiple PiggyBac Vectors in a single transfection
- Insert an expression cassette into human, mouse, and rat cells
- Deliver virtually any-sized DNA insert, from 10 – 100 kb
- Choose from PiggyBac Vectors that express your gene-of-interest from constitutive or inducible promoters and include a variety of markers
- Determine the number of integration events with the PiggyBac qPCR Copy Number Kit (# PBC100A-1)
Customer Agreements
Academic customers can purchase PiggyBac Transposon System components for internal research purposes for indefinite use, whereas commercial customers must sign a customer agreement for a six-month, limited-use license to evaluate the technology.
For end user license information, see the following:
* SBI is fully licensed to distribute PiggyBac vectors as a partnership with Hera BioLabs, Inc.
How It Works
How It Works
The PiggyBac Transposon System’s Cut-and-Paste Mechanism
The efficient PiggyBac Transposon System uses a cut-and-paste mechanism to transfer DNA from the PiggyBac Vector into the genome. If only temporary genomic integration is desired, the Excision-only PiggyBac Transposase can be transiently expressed for footprint-free removal of the insert, resulting in reconstitution of the original genome sequence.
Figure 1. The PiggyBac Transposon System’s cut-and-paste mechanism.
- The Super PiggyBac Transposase binds to specific inverted terminal repeats (ITRs) in the PiggyBac Cloning and Expression Vector and excises the ITRs and intervening DNA.
- The Super PiggyBac Transposase inserts the ITR-Expression Cassette-ITR segment into the genome at TTAA sites.
- The Excision-only Super PiggyBac Transposase can be used to remove the ITR-Expression Cassette-ITR segment from the genome, for footprint-free removal
Supporting Data
FAQs
Citations
-
Brouwer, I, de Kort, MAC & Lenstra, TL. (2024) Measuring Transcription Dynamics of Individual Genes Inside Living Cells. Methods in molecular biology (Clifton, N.J.). 2024; 2694:235-265. PM ID: 37824008
-
Matta, SK, et al. (2024) Genome-wide and targeted CRISPR screens identify RNF213 as a mediator of interferon gamma-dependent pathogen restriction in human cells. Proceedings of the National Academy of Sciences of the United States of America. 2024; 121(1):e2315865120. PM ID: 38147552
-
Cho, MG, et al. (2024) MRE11 liberates cGAS from nucleosome sequestration during tumorigenesis. Nature. 2024; 625(7995):585-592. PM ID: 38200309
-
Du, M, et al. (2024) Direct observation of a condensate effect on super-enhancer controlled gene bursting. Cell. 2024; 187(2):331-344.e17. PM ID: 38194964
-
Schmitt, J, et al. (2024) Repurposing an endogenous degradation domain for antibody-mediated disposal of cell-surface proteins. EMBO reports. 2024;. PM ID: 38287192
-
Byrnes, AE, et al. (2024) A fluorescent splice-switching mouse model enables high-throughput, sensitive quantification of antisense oligonucleotide delivery and activity. Cell reports methods. 2024; 4(1):100673. PM ID: 38171361
-
Daiki, K, et al. (2024) Blood Endocan as a Biomarker for Breast Cancer Recurrence. Preprint. 2024;. Link: Preprint
-
Koeppel, J, et al. (2024) Randomizing the human genome by engineering recombination between repeat elements. bioRxiv. 2024;. Link: bioRxiv
-
Kortleve, D, et al. (2024) TCR-engineered T-cells directed against Ropporin-1 constitute a safe and effective treatment for triple-negative breast cancer in near-clinical models. bioRxiv. 2024;. Link: bioRxiv
-
Haakonsen, DL, et al. (2024) Stress response silencing by an E3 ligase mutated in neurodegeneration. Nature. 2024; 626(8000):874-880. PM ID: 38297121
-
Gupta, P, et al. (2024) Development of pathophysiologically relevant models of sickle cell disease and β-thalassemia for therapeutic studies. Nature communications. 2024; 15(1):1794. PM ID: 38413594
-
Company, C, et al. (2024) Logical design of synthetic cis-regulatory DNA for genetic tracing of cell identities and state changes. Nature communications. 2024; 15(1):897. PM ID: 38316783
-
Yang, L, et al. (2024) Uncovering receptor-ligand interactions using a high-avidity CRISPR activation screening platform. Science advances. 2024; 10(7):eadj2445. PM ID: 38354234
-
Kubara, K, et al. (2024) Lymph node macrophages drive innate immune responses to enhance the anti-tumor efficacy of mRNA vaccines. Molecular therapy : the journal of the American Society of Gene Therapy. 2024;. PM ID: 38243602
-
Ng-Blichfeldt, J, et al. (2024) Identification of a core transcriptional program driving the human renal mesenchymal-to-epithelial transition. Developmental Cell. 2024;. Link: Developmental Cell
-
Yang, J, Cook, L & Chen, Z. (2024) Systematic evaluation of retroviral LTRs as cis-regulatory elements in mouse embryos. Cell reports. 2024; 43(3):113775. PM ID: 38381606
-
Taglini, F, et al. (2024) DNMT3B PWWP mutations cause hypermethylation of heterochromatin. EMBO reports. 2024;. PM ID: 38291337
-
Tanase-Nakao, K, et al. (2024) Genotype-Phenotype Correlations in Thirty Japanese Patients with Congenital Hypothyroidism Attributable to TG Defects. The Journal of clinical endocrinology and metabolism. 2024;. PM ID: 38373250
-
Alsouri, S, et al. (2024) Actinin-4 controls survival signaling in B cells by limiting the lateral mobility of B-cell antigen receptors. European journal of immunology. 2024;:e2350774. PM ID: 38299456
-
Ke, X, et al. (2024) Establishment of a novel minigenome system for the identification of drugs targeting Nipah virus replication. The Journal of general virology. 2024; 105(1). PM ID: 38180473
- See More